Abstract
Introduction
This study aims to construct a mortality prediction model for patients with non-variceal upper gastrointestinal bleeding (NVUGIB) in the intensive care unit (ICU), employing advanced machine learning algorithms. The goal is to identify high-risk populations early, contributing to a deeper understanding of patients with NVUGIB in the ICU.
Methods
We extracted NVUGIB data from the Medical Information Mart for Intensive Care IV (MIMIC-IV, v.2.2) database spanning from 2008 to 2019. Feature selection was conducted through LASSO regression, followed by training models using 11 machine learning methods. The best model was chosen based on the area under the curve (AUC). Subsequently, Shapley additive explanations (SHAP) was employed to elucidate how each factor influenced the model. Finally, a case was randomly selected, and the model was utilized to predict its mortality, demonstrating the practical application of the developed model.
Results
In total, 2716 patients with NVUGIB were deemed eligible for participation. Following selection, 30 out of a total of 64 clinical parameters collected on day 1 after ICU admission remained associated with prognosis and were utilized for developing machine learning models. Among the 11 constructed models, the Gradient Boosting Decision Tree (GBDT) model demonstrated the best performance, achieving an AUC of 0.853 and an accuracy of 0.839 in the validation cohort. Feature importance analysis highlighted that shock, Glasgow Coma Scale (GCS), renal disease, age, albumin, and alanine aminotransferase (ALP) were the top six features of the GBDT model with the most significant impact. Furthermore, SHAP force analysis illustrated how the constructed model visualized the individualized prediction of death.
Conclusions
Patient data from the MIMIC database were leveraged to develop a robust prognostic model for patients with NVUGIB in the ICU. The analysis using SHAP also assisted clinicians in gaining a deeper understanding of the disease.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
Non-variceal upper gastrointestinal bleeding (NVUGIB) refers to bleeding that develops in the esophagus, stomach, or proximal duodenum. The annual incidence of NVUGIB ranges from 19.4 to 57.0 cases per 100,000 in the population, accompanied by a mortality rate of approximately 6–12%. This condition incurs an annual hospital cost exceeding $2.5 billion in the United States and is a common presentation in intensive care unit (ICU) worldwide [1,2,3]. Therefore, early risk stratification of patients with NVUGIB is imperative for the identification of high- and low-risk individuals.
Various risk scores have been devised to anticipate the mortality associated with NVUGIB. In an international multicenter prospective study, a comparison was made among the Glasgow Blatchford score (GBS), Rockall score, Progetto Nazionale Emorragia Digestive (PNED) score, and AIMS65 score (AIM). The study revealed that GBS emerged as the most reliable predictor of the need for intervention, while the AIMS65 score demonstrated the highest accuracy in predicting mortality [4]. Moreover, numerous clinical trials have demonstrated the substantial impact of factors such as coagulation, comorbidities, and low albumin levels on the prognosis of gastrointestinal bleeding [5, 6]. Simultaneously, various studies have endeavored to formulate scoring systems tailored to different patient populations experiencing gastrointestinal bleeding [7,8,9]. Nevertheless, as of now, no predictive model has been specifically crafted for application in the ICU.
Machine learning (ML) endeavors to replicate human abilities in recognizing patterns, constructing predictive models from data, and discerning meaningful data groupings [10]. In divergence from traditional predictive models, which depend on selected variables for calculations, machine learning techniques exhibit the capacity to seamlessly integrate a vast number of variables, given that all computations are executed by a computer [11]. ML algorithm utilized for mortality prediction facilitates impartial processing of extensive clinical variables, allowing for the identification of crucial factors in a non-supervised manner. These algorithms empower the identification of distinct patient phenotypes and enable the visualization of the quantitative contribution of each variable to the outcome [12]. Nevertheless, the 'black-box' nature inherent in ML algorithms poses challenges when it comes to elucidating the rationale behind specific predictions concerning patients or the particular patient characteristics influencing those predictions. To mitigate these challenges, this study integrates advanced ML algorithms with a framework founded on Shapley additive explanations (SHAP) [13]. SHAP offers intuitive explanations, enhancing the comprehensibility of risk predictions for patients.
The objective of this study is to formulate multiple predictive models utilizing machine learning techniques and identify the optimal-performing model. Following model selection, we will employ SHAP to interpret the chosen model, thereby unveiling the significant influential factors for NVUGIB in the ICU.
Methods
Study design
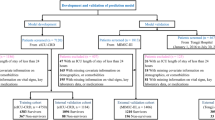
In this retrospective study, patient data were extracted from the Medical Information Mart for Intensive Care IV (MIMIC-IV, v.2.2) database. Subsequently, ten distinct machine learning methods were applied to construct a predictive model for mortality. The model that exhibited superior performance was chosen and further interpreted using the SHAP method.
Data
We collected data for this study from the Medical Information Mart for Intensive Care IV (MIMIC-IV, v.2.2) database. MIMIC-IV is a large, freely accessible database containing clinical data from adult patients (aged 18 years and older) admitted to an ICU at a major tertiary care hospital in the US between 2008 and 2019. It encompasses over 70,000 ICU admissions [14]. One of our team members has effectively fulfilled the Collaborative Institutional Training Initiative (CITI) examination (Record ID: 47937607 for CH) and has been granted authorization to access the MIMIC-IV database. Approval for our project has been secured from the Institutional Review Boards of Beth Israel Deaconess Medical Center (Boston, MA, USA) and the Massachusetts Institute of Technology (Cambridge, MA, USA). The project was approved by the institutional review boards of the Massachusetts Institute of Technology (MIT) and Beth Israel Deaconess Medical Center (BIDMC) and was granted a waiver of informed content. Additionally, our study adheres to the principles of the Declaration of Helsinki and follows the guidelines for Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis.
Participants
All patients presenting with NVUGIB were included in our study from the MIMIC-IV database. The diagnostic ICD codes are detailed in Table 1. Exclusion criteria for study participation comprised: (1) patients with multiple ICU admissions (only the initial admission was considered for analysis), and (2) patients with an ICU length of stay less than 24 h. The primary clinical outcome was in-hospital mortality.
Variable selection
We meticulously selected 64 candidate variables based on their association with the outcome. These encompassed demographic factors such as age, gender, body weight, and height; comorbidities including cerebrovascular disease, heart failure, hypertension, shock, chronic pulmonary disease, liver disease, renal disease, tumor, diabetes, hepatic failure, melena, and syncope; vital signs (mean values in the ICU) such as heart rate, systolic blood pressure, diastolic blood pressure, mean arterial pressure, respiratory rate, and body temperature; laboratory findings (maximum values) encompassing blood glucose, lactate, pH, PCO2, white blood cell count, anion gap, bicarbonate, blood urea nitrogen, serum chloride, serum creatinine, serum sodium, serum fibrinogen, serum potassium, international normalized ratio, prothrombin time, partial thromboplastin time, alanine aminotransferase (ALT), alkaline phosphatase (ALP), aspartate aminotransferase (AST), total bilirubin, amylase, creatine phosphokinase, creatine kinase MB, and lactate dehydrogenase; as well as the minimum values for SPO2, PO2, PaO2/FiO2 ratio, hematocrit, hemoglobin, platelets, and albumin. Additionally, we incorporated the Glasgow Coma Scale (GCS) and AIMS65 (albumin level < 30 g/L (A), international normalized ratio > 1.5 (I), altered mental status (M), systolic blood pressure ≤ 90 mm Hg (S), and age > 65 years (65)) score (Table S1).
For each aforementioned variable, we extracted the value recorded within the initial 24 h of ICU admission. Features exhibiting more than 30% missing data were excluded, as illustrated in Supplementary Figure S2. To address missing values in the remaining features, we applied multiple imputation by chained equations (MICE). Given the substantial number of features in the cohort, variable selection was executed using the least absolute shrinkage and selection operator (LASSO) regression, a method adept at preventing overfitting. The comprehensive list of input variables utilized in the model development is available in Supplementary Table S2.
Statistical analysis
Categorical data are presented as counts and percentages, and between-group differences were evaluated using the Chi-squared test or Fisher's exact test. Continuous data are expressed as means with standard deviations (SDs) or medians with interquartile ranges (IQRs), depending on the distribution of the data. Group differences for continuous data were analyzed using either analysis of variance or the Mann–Whitney U test.
To address missing data with less than 30% in each feature, we utilized multiple imputation via the 'mice' package in R. Feature selection was executed through LASSO regression. The population was randomly divided into two segments, with 80% designated as the derivation data and 20% as the validation data. Our feature engineering process for the variables includes two main steps: outlier processing and normalization. Firstly, we employ a robust similarity measure based on the absolute deviation of the median to handle outliers. This approach ensures that our datasets are more resilient and dependable during the modeling process. Secondly, we normalize the continuous variables by converting them to the Z-scal, resulting in a mean of 0 and a standard deviation of 1. After tuning hyperparameters for 11 common machine learning methods, including Support Vector Machine (SVM), k-Nearest Neighbors (KNN), eXtreme Gradient Boosting (XGBoost), Decision Tree (DT), Naive Bayes (NB), Random Forest (RF), Adaptive Boosting (AdaBoost), Gradient Boosting Decision Tree (GBDT), Multilayer Perceptron (MLP), Light Gradient Boosting Machine (LGBM), Catboost, and logistic regression (LR), models were developed in the training cohort. The hyperparameters of each model were tuned using the GridSearchCV, which employs grid search and cross-validation techniques. The tuned hyperparameters for each model are specified in Table S3.
Performance evaluation of the models in predicting mortality was conducted using the area under the curve (AUC) and accuracy metrics in the validation cohort. The model exhibiting the best performance in the validation cohort was designated as the final model. Additionally, we assessed the predictive capabilities of illness severity scoring systems, including the Glasgow Blatchford Score (GBS) and AIMS65 score, to predict the risk of death and compared their performance with our final model in the validation cohort.
Furthermore, we employed the SHAP method to augment the interpretability of the ultimate model. Initially, the SHAP summary plot was utilized to depict the effects of features ascribed to the model. Subsequently, the SHAP dependence plot was employed to scrutinize the individual influence of features on the model's output. Finally, the SHAP force plot was applied to visually represent the impact of pivotal features on the ultimate model for individual patients.
Based on the results provided by SHAP, we further explored the interaction between the first six variables using Spearman correlation. Subsequently, we conducted interaction analysis for variables exhibiting significant correlations.
Simultaneously, to comprehensively understand patient prognosis, we extracted the length of hospital stay, 90-day mortality, and 1-year mortality as secondary outcome indicators. For 90-day mortality and 1-year mortality, we applied the same methods used previously to fit machine learning models, followed by SHAP analysis to explain the best-performing model. For length of stay, we utilized Cox proportional hazards model (COX) regression to assess the impact of each factor on the length of stay and outcomes.
All analyses were conducted using Python (v.3.6.6) and R (v. 3.6.1, R Foundation for Statistical Computing). Significance was determined by two-tailed P values < 0.05.
Sensitivity analysis
We conducted a sensitivity analysis by employing various methods for screening variables. Specifically, we utilized the Recursive Feature Elimination (RFE) method to extract features. This method begins with a complete feature set based on specific criteria for feature ordering, then iteratively removes the least relevant features until the most important ones are identified. Subsequently, we selected the top 30 most important features and trained the GBDT model, which was further elucidated using SHAP.
Results
Participants
Among the 76,540 critically ill patients evaluated from the MIMIC-IV database, a screening process identified 9191 individuals with NVUGIB. Following the exclusion of 5741 patients with multiple ICU admissions (considering only the first ICU admission for analysis) and 734 patients with an ICU stay of less than 24 h, a cohort of 2716 patients met the eligibility criteria for participation (refer to Supplementary Figure S1).
Baseline characteristics of the eligible participants are summarized in Table 1. The median age was 66.0 years (IQR, 54–77.75 years), with 1046 out of 2716 participants (38.5%) being women. Renal disease was the most common comorbidity (1598 out of 2716, 58.84%), followed by liver disease (1111 out of 2716, 40.91%), and hypertension (983 out of 2716, 36.19%).
Model development and validation
We collected a total of 64 clinical variables during the first 24 h after ICU admission. Among them, 10 variables with over 30% missing data were excluded (Supplementary Figure S2). Missing data in the remaining feature space were imputed using MICE. LASSO regression was used to identify signature variables for hospital mortality in patients with NVUGIB, resulting in 30 significant clinical parameters (Supplementary Figure S3 and Table S2). We constructed 11 machine learning binary classifiers to predict the risk of death in NVUGIB patients: Support Vector Machine (SVM), k-Nearest Neighbors (KNN), eXtreme Gradient Boosting (XGB), Decision Tree (DT), Random Forest (RF), Adaptive Boosting (AdaBoost), Gradient Boosting Decision Tree (GBDT), Multilayer Perceptron (MLP), Light Gradient Boosting Machine (LGBM), Catboost, and logistic regression (LR) (Fig. 1). The performance of these predictive models is described in Fig. 2A, with GBDT showing the best model fit performance, achieving an AUC of 0.853 and an accuracy of 0.839 in the validation cohort compared to the other ML models (AUC: KNN, 0.818; DT, 0.760; RF, 0.849; SVM, 0.831; XGB, 0.850; AdaBoost, 0.834; MLP, 0.820; LGBM, 0.849; Cat, 0.834; Logistic, 0.837. Accuracy: KNN, 0.827; DT, 0.809; RF, 0.836; SVM, 0.827; XGB, 0.830; AdaBoost, 0.843; MLP, 0.836; LGBM, 0.838; Cat, 0.836; Logistic, 0.835) (Table 2). Figure 2B displays the AUCs of the analyzed scores and compares them to each other. The GBDT model outperformed other severity of illness scores (AUC: GBDT, 0.853; AIMS65, 0.644; GBS, 0.551). The calibration curve for the GBDT model indicates good agreement between predicted and actual risk (Supplementary Figure S4). Consequently, GBDT was chosen for further prediction in this study.
Receiver operator characteristic (ROC) curves for the ML models and the traditional severity of illness scores to predict in-hospital mortality (validation cohort). A ROC curves for the seven ML models to predict in-hospital mortality; B ROC curves for the traditional severity of illness scores to predict in-hospital mortality. Support Vector Machine (SVM), k-Nearest Neighbors (KNN), eXtreme Gradient Boosting (XGBoost), Decision Tree (DT), Naive Bayes (NB), Random Forest (RF), Adaptive Boosting (AdaBoost), Gradient Boosting Decision Tree (GBDT), Multilayer Perceptron (MLP), Light Gradient Boosting Machine (LGBM), Catboost (Cat), and logistic regression (LR), Glasgow Blatchford Score (GBS) and AIMS65 score (AIM)
Model explanation
We computed feature importance using SHAP values for the GBDT model, which exhibited the highest discriminatory ability in the validation cohort. Figure 3A illustrates the top 20 clinical features based on the average absolute SHAP values. Figure 3B offers an overview of the positive or negative impact of factors on the GBDT model. Features specific to death included a lower Glasgow Coma Scale (GCS) score, albumin, serum chloride, SPO2, and weight, each with a negative SHAP value, driving predictions toward mortality. Elevated age, ALP, respiratory rate, lactate, PT, and total bilirubin also supported a prediction of mortality. The impact of the top six factors on the GBDT model's mortality risk prediction was further explored using SHAP dependency plots. Figure 4 illustrates that higher levels of ALP, respiratory rate, older age, lower GCS, lower albumin, and lower serum chloride were associated with a significant increase in mortality risk.
SHAP summary plot for the top 20 clinical features contributing to the GBDT model. A SHAP feature importance measured as the mean absolute Shapley values. This matrix plot depicts the importance of each covariate in the development of the final predictive model. B The attributes of the features in the model. The position on the y-axis is determined by the feature and on the x-axis by the Shapley value. The color represents the value of the feature from low to high
Model application
To further investigate the contribution of features to an individual patient and their clinical application for the GBDT model, we randomly selected one patient from the validation cohort. Figure 5 offers a visual interpretation of the GBDT model. The model predicted a 51% probability of death for this patient (Fig. 5A). The top three contributors to this prediction were identified as ALP with a value of 941, GCS score of 10, and the absence of a shock diagnosis. Figure 5B illustrates the specific impact of these factors on the prediction.
SHAP force plot for explaining of individual’s prediction results in the validation cohort. Screenshot of the death prediction in patients with NVUGIB. A, B Model predictions by randomly drawing a single sample from the validation cohort. Redder sample points indicate that the value of the feature is larger, and bluer sample points indicate that the value of the feature is smaller
Sensitivity analysis
We utilized the RFE method to re-screen 30 characteristic variables. Among these, 17 variables were identified in the LASSO regression screening, while 13 were not. The specific variables are detailed in Table S4. The ROC curve subsequent to fitting the GBDT model twice is illustrated in Figure S5. Following SHAP analysis, the importance ranking of each variable in the model is presented in Figure S5. Despite large changes in the included variables, SHAP analysis revealed that the first six variables, which had the most impact on the model, remained unchanged.
Interaction analysis
Based on the GBDT fitting model and sensitivity analysis results, we selected the first six variables that had the greatest impact on the model in the two experiments. We then applied Spearman correlation analysis to these variables, and the results are presented in Figure S6A. Subsequently, we conducted interaction analysis on variables exhibiting significant correlations, and the results are shown in Figure S6B-L.
Secondary outcome assessment
The model fitting results, with 90-day survival rate and 1-year survival rate as outcomes, are depicted in Figure S7. The findings reveal that when using 90-day mortality as the outcome indicator, the random forest fitting model exhibited the highest AUC of 0.789, with shock, age, ALP, GCS and albumin identified as the five factors exerting the greatest impact on the model. Conversely, for 1-year mortality as the outcome, the random forest model demonstrated optimal fitting (AUC = 0.773), highlighting age, ALP, shock, kidney disease and GCS as the most influential factors. The results of multivariate Cox regression analysis on length of stay are summarized in Table S5, and the Cox regression curve is depicted in Figure S8.
Discussion
Non-variceal upper gastrointestinal bleeding (NVUGIB) is a common condition in the ICU, yet there was no appropriate model available for predicting NVUGIB mortality in this setting. While AIMS65 and GBS performed well in some studies, our research showed poor performance for these scores (AIMS65 AUC = 0.664, GBS AUC = 0.551). This indicates the need for separate prediction models for ICU patients. We leveraged machine learning methods, employing 11 common ML techniques to develop a prediction model for NVUGIB in the ICU. The GBDT model achieved the highest AUC score. Feature importance analysis identified shock, GCS, renal disease, age, albumin, and ALP as the top six predictors of mortality. We explored how these features influenced the GBDT model, and provided an example demonstrating how the GBDT model predicts NVUGIB mortality using SHAP force analysis. Our study contributes a mortality prediction model for NVUGIB patients in the ICU, enhancing our understanding of this condition.
We utilized SHAP to interpret the GBDT model and unveil its 'black box'. Developed by Lundberg and Lee, SHAP is a mathematical framework used in machine learning for explaining the output of complex models. It is rooted in cooperative game theory and provides a way to fairly distribute contributions of individual features to the prediction made by a model [15]. It has seen extensive use in explaining ML models in the medical field [16]. Our SHAP analysis revealed that shock, GCS score, renal disease, age, albumin, and ALP were the six most influential factors in predicting mortality in our study.
Shock emerges as the most critical factor in our model. Massive hemorrhage in NVUGIB patients can lead to shock, resulting in organ hypoperfusion and impaired organ function. But for NVUGIB patients, estimating the amount of bleeding can be challenging, and our study's results reinforce that shock has the most significant impact on patient mortality. Therefore, early diagnosis and confirmation of shock are paramount. In the GBS and AIMS65 systems, systolic blood pressure was considered [17, 18]. Although we included systolic blood pressure, diastolic blood pressure and mean arterial pressure in our model, LASSO regression excluded these factors. This suggests that shock can provide a more accurate prognosis evaluation than these blood pressure-related factors in this patient group.
Similar to the AIMS65, our model incorporated age. Multicenter studies have also indicated a higher risk of death in elderly patients [19, 20]. Our study reinforces the association between aging and elevated mortality. Given the multifactorial nature of poor prognosis in elderly patients with gastrointestinal bleeding, it underscores the necessity for more meticulous treatment plans for middle-aged and elderly patients who often face a worse prognosis.
GCS reflects patients' neurological function and, to some extent, the rate and total amount of bleeding. Our findings highlight GCS as the second most critical prognostic factor after shock. Lower GCS scores were associated with higher mortality, possibly due to acute gastrointestinal bleeding causing hemodynamic instability, leading to neurologic symptoms. A lower GCS score is indicative of less hemodynamic stability and increased blood loss [21]. GCS score can be utilized for the identification not only of high-risk cases, but also in discerning underlying neurological diseases. This underscores the dual utility of the GCS score in not just assessing risk, but also aiding in the determination of the root cause [22].
Our experiment revealed a significant association between albumin levels and mortality in patients with NVUGIB. Univariate analysis demonstrated that a decrease in albumin was correlated with an increase in mortality. The reduction in albumin levels may be attributed to substantial blood loss. Furthermore, low albumin levels may also be indicative of underlying conditions such as chronic liver disease, kidney disease, malnutrition, and other contributing factors [23], all of which may be implicated in gastrointestinal bleeding. In conclusion, monitoring the albumin levels of patients upon admission is crucial. This practice aids in assessing the severity and determining the etiology of the disease.
Renal disease emerged as the third most significant feature in our model. Alongside anemia, coagulation disorders constitute major hematological abnormalities observed in renal pathology. The accumulation of uremic toxins in patients with end-stage renal disease promotes platelet abnormalities, thereby contributing to thrombotic events. Numerous additional risk factors, including blood abnormalities, inflammation, comorbidities, and endothelial dysfunction, may further predispose individuals to such events [24]. In our study, platelets, INR, PT, and PTT were utilized to construct the model. Notably, PT emerged as relatively important, ranking twelfth in significance. Despite the inclusion of coagulation measures, they exhibited a lesser impact on the model compared to renal disease. This suggests that renal disease may contribute to impaired coagulation, while concurrently serving as a more effective predictor of patient mortality. Consequently, NVUGIB patients presenting to the ICU with renal disease warrant particular attention. Further prospective randomized controlled trials are imperative to delve into the intricate relationship between kidney disease, coagulation, and gastrointestinal bleeding.
In conclusion, our developed model for NVUGIB in the ICU reveals that NVUGIB patients with shock, renal disease, low GCS scores, low albumin levels, and advanced age have higher mortality rates. Physicians should prioritize the care of these patients and closely monitor their condition. Moreover, as depicted in Fig. 5, specific laboratory findings or vital signs such as extreme ALP levels should also warrant attention. Additionally, according to Figure S6B, simultaneous occurrence of shock and renal disease in patients necessitates immediate intervention. Hemorrhagic shock can decrease renal perfusion and lead to acute kidney injury (AKI), with renal disease potentially developing as a secondary consequence of shock and serving as an indicator of the severity of shock [25]. Meanwhile, renal disease can impair coagulation, acting as a primary predisposing condition in the pathogenesis of NVUGIB [26]. When managing NVUGIB patients presenting with shock and renal disease, early and accurate identification of etiology and tailored, individualized treatment strategies are crucial.
Several limitations should be considered in our study. Firstly, it was a single-center retrospective modeling study using the MIMIC-IV database, limiting our ability to establish causal relationships between features and outcomes. Prospective randomized clinical trials are needed for further validation. Secondly, unmeasured confounders, including racial and treatment differences, may impact the prognosis of NVUGIB patients. Therefore, some crucial features for the GBDT model might have been overlooked due to the limitations of the MIMIC-IV database. Thirdly, imputation and feature selection were executed prior to the division of the dataset into training and validation sets, introducing a potential impact on the ultimate models. Fourthly, our study is characterized by a deficiency in external validation, as it lacks an independent cohort from different regions or countries, underscoring the imperative for additional investigations to substantiate our findings. Lastly, it is noteworthy that our modeling study exclusively encompassed adults, leaving the predictive efficacy of the GBDT model for pediatric patients with NVUGIB uncertain. Therefore, prudent interpretation of our conclusions is advised, emphasizing the requisite for additional empirical support in subsequent research endeavors.
Availability of data and materials
No datasets were generated or analysed during the current study.
References
Seo DW, et al. Prediction of adverse events in stable non-variceal gastrointestinal bleeding using machine learning. J Clin Med. 2020;9(8):2603.
Kate V, et al. Acute upper non-variceal and lower gastrointestinal bleeding. J Gastrointest Surg. 2022;26(4):932–49.
Lanas A, et al. Non-variceal upper gastrointestinal bleeding. Nat Rev Dis Primers. 2018;4:18020.
Stanley AJ, et al. Comparison of risk scoring systems for patients presenting with upper gastrointestinal bleeding: international multicentre prospective study. BMJ. 2017;356: i6432.
El HW, et al. Prognosis of variceal and non-variceal upper gastrointestinal bleeding in already hospitalised patients: Results from a French prospective cohort. United European Gastroenterol J. 2021;9(6):707–17.
Kim WS, et al. Re-bleeding and all-cause mortality risk in non-variceal upper gastrointestinal bleeding: focusing on patients receiving oral anticoagulant therapy. Ann Med. 2023;55(2):2253822.
Quach DT, et al. Development and validation of a scoring system to predict severe acute lower gastrointestinal bleeding in Vietnamese. Dig Dis Sci. 2021;66(3):823–31.
Ito N, et al. A novel scoring system to predict therapeutic intervention for non-variceal upper gastrointestinal bleeding. Intern Emerg Med. 2022;17(2):423–30.
Zhang X, et al. A nomogram to predict in-hospital mortality of gastrointestinal bleeding patients in the intensive care unit. Front Med. 2023;10:1204099.
Zhang X, et al. The predictive value of machine learning for mortality risk in patients with acute coronary syndromes: a systematic review and meta-analysis. Eur J Med Res. 2023;28(1):451.
Nistal-Nuno B. Developing machine learning models for prediction of mortality in the medical intensive care unit. Comput Methods Programs Biomed. 2022;216: 106663.
Barghi B, Azadeh-Fard N. Predicting risk of sepsis, comparison between machine learning methods: a case study of a Virginia hospital. Eur J Med Res. 2022;27(1):213.
Zhou S, et al. Interpretable machine learning model for early prediction of 28-day mortality in ICU patients with sepsis-induced coagulopathy: development and validation. Eur J Med Res. 2024;29(1):14.
Johnson, A., et al., MIMIC-IV (version 2.2). PhysioNet. https://doi.org/10.13026/6mm1-ek67. 2021.
Junaid M, et al. Explainable machine learning models based on multimodal time-series data for the early detection of Parkinson’s disease. Comput Methods Programs Biomed. 2023;234: 107495.
Zhang G, et al. Predicting sepsis in-hospital mortality with machine learning: a multi-center study using clinical and inflammatory biomarkers. Eur J Med Res. 2024;29(1):156.
Blatchford O, Murray WR, Blatchford M. A risk score to predict need for treatment for upper-gastrointestinal haemorrhage. Lancet. 2000;356(9238):1318–21.
Saltzman JR, et al. A simple risk score accurately predicts in-hospital mortality, length of stay, and cost in acute upper GI bleeding. Gastrointest Endosc. 2011;74(6):1215–24.
Wong M, et al. Differences in incidence and mortality trends of colorectal cancer worldwide based on sex, age, and anatomic location. Clin Gastroenterol Hepatol. 2021;19(5):955-966.e61.
Zhang JJ, et al. Clinical, radiological, and laboratory characteristics and risk factors for severity and mortality of 289 hospitalized COVID-19 patients. Allergy. 2021;76(2):533–50.
Kaya E, et al. Predictors of poor outcome in gastrointestinal bleeding in emergency department. World J Gastroenterol. 2016;22(16):4219–25.
Qiu W, et al. Age-to-Glasgow Coma Scale score ratio predicts gastrointestinal bleeding in patients with primary intracerebral hemorrhage. Front Neurol. 2023;14:1034865.
Manolis AA, et al. Low serum albumin: a neglected predictor in patients with cardiovascular disease. Eur J Intern Med. 2022;102:24–39.
Pavlou EG, et al. Coagulation abnormalities in renal pathology of chronic kidney disease: the interplay between blood cells and soluble factors. Biomolecules. 2021;11(9):1309.
Messerer D, et al. Immunopathophysiology of trauma-related acute kidney injury. Nat Rev Nephrol. 2021;17(2):91–111.
Garg R, et al. Lower rates of endoscopy and higher mortality in end-stage renal disease patients with gastrointestinal bleeding: a propensity matched national study. J Gastroenterol Hepatol. 2022;37(3):584–91.
Acknowledgements
We appreciate Dr. Shuxing Wei from the Beijing Key Laboratory of Cardiopulmonary Cerebral Resuscitation, for the suggestions regarding the statistical analysis and manuscript preparation.
Funding
There was no funding for this study.
Author information
Authors and Affiliations
Contributions
X. Z., S. W. and Y. P. contributed to the statistical analysis and mathematical modeling. K. Q., G. Y., X. W. and Y. S. contributed to the scientific oversight and literature analysis. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was an analysis of a third party anonymized publicly available database with pre-existing institutional review board (IRB) approval.
Consent for publication
My co-authors have all contributed to approve of this submission.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
40001_2024_2005_MOESM1_ESM.docx
Supplementary Material 1: Figure S1. Study flowchart. Figure S2. Percentages of missing data for all included variables. Figure S3. Feature selection using the LASSO regression model. (A) LASSO coefficient profiles of the 47 baseline features. (B) Tuning parameterselection in the LASSO model used fivefold cross-validation via minimum criteria. LASSO, least absolute shrinkage and selection operator. Figure S4. Calibration curve for in-hospital mortality for validation cohort in GBDT. The dotted line indicates the ideal reference line where predicted probabilities would match the observed survival rates. The blue line represents the performance of the GBDT. The closer the blue line is to the dotted line, the more accurately the model predicts in-hospital mortality. Figure S5. Sensitivity analysis result. A: ROC curves for the traditional severity of illness scores to predict in-hospital mortality and sensitivity analysis. B: SHAP feature importance measured as the mean absolute Shapley values. This matrix plot depicts the importance of each covariate in the development of the final predictive model. C: The attributes of the features in the model. The position on the y-axis is determined by the feature and on the x-axis by the Shapley value. The color represents the value of the feature from low to high. Figure S6. Sensitivity analysis result. A: Spearman analysis results of correlation between variables. Numbers indicate the P value of the correlation analysis, circles indicate the correlation coefficient B-L: Analysis results of the interaction between correlated variables. Figure S7. Results of analyses of secondary outcomes. A–C: Machine learning model fit and SHAP interpretation with 30-day mortality as the outcome. D–F: Machine learning model fit and SHAP interpretation with 1 year mortality as the outcome. Figure S8. COX regression curve of the length of hospital stay. Table S1. All extracted variables in MIMIC-IV database. Table S2. Clinical features used for developing the models. Table S3. The tuned hyperparameters for the eleven models.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, X., Wei, S., Pan, Y. et al. Early prognosis prediction for non-variceal upper gastrointestinal bleeding in the intensive care unit: based on interpretable machine learning. Eur J Med Res 29, 442 (2024). https://doi.org/10.1186/s40001-024-02005-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40001-024-02005-0