Abstract
Background
Diarrhoea is a major cause of childhood disease in the developing countries. This experimental study investigated the prevalence of ESBL and MBL genes in enteropathogenic strains of Escherichia coli and Salmonella spp. isolated from diarrheagenic children in Awka, Nigeria.
Methods
Two hundred stool samples were collected from diarrhea patients in three paediatric hospitals within Awka metropolis, Nigeria. All E. coli and Salmonella spp. isolated through standard bacteriological methods were subjected to antibiotic-susceptibility testing. Double disc synergy and imipenem-EDTA combined disc tests were used to phenotypically confirm the presence of ESBL and MBL respectively. PCR amplification of β-lactamase genes was done.
Results
The prevalence of E. coli and Salmonella species in this study were 54% and 24.5% respectively. The organisms were highly resistant to metronidazole, cefuroxime and ceftazidime, and also showed a high sensitivity to nitrofurantoin and gentamicin. ESBL production was recorded in E. coli (49%) and Salmonella spp. (51.1%) while 27 isolates of E. coli (25%) and 7 isolates of Salmonella spp. were confirmed MBL positive by the combined disk diffusion technique. Eleven E. coli and 4 Salmonella spp. co-harbored both ESBL and MBL production. The most prevalent MBL gene in this study is the blaVIM gene (18.8%) which mediate MBL production in Gram negative bacteria; and this was followed by blaSHV (12.5%), blaTEM and blaCTX-M (6.3% each) for E. coli isolates. Salmonella spp. was recorded to have blaVIM (28.8%), blaSHV (28.8%), blaTEM (14.3%) and blaCTX-M (14.3%) genes.
Conclusions
This study reveals the prevalence of enteropathogenic E. coli and Salmonella strains bacteriologically recovered from diarrheic children in Awka, Nigeria, and which were found to be multiple resistant to clinically-relevant antibiotics because they co-express ESBL and MBL genes which mediate multidrug resistance in Gram negative bacteria.
Similar content being viewed by others
Background
Diarrhoeal disease is the most common health problem in developing countries especially in Africa and the leading cause of hospitalization and sometimes even death among young children (Ugboko et al. 2020; Ugwu et al. 2017). In Nigeria, it has been estimated that approximately 10.12% of child deaths during the first five years of life are associated with diarrhoeal diseases (WHO 2018). Diarrhoea accounts for nine percent of all deaths and pediatric admission among children under age five worldwide (Tsehay et al. 2021). Diarrhoea infections claimed about 580,000 children’ under age five yearly or on average of 1600 children daily despite the availability of treatment options (WHO 2018). Escherichia coli and Salmonella species are the common bacterial etiological agent associated with childhood diarrhea (WHO 2018; Okoli et al. 2021). Both are enteropathogenic, Gram-negative facultative anaerobe which belongs to the family Enterobacteriaceae (Adesoji and Laidi 2020). Escherichia coli and Salmonella spp are mostly isolated food-borne pathogens predominantly found in animal products, fresh fruits and vegetables; and ready to eat foods (Ugwu et al. 2020, 2019; Heredia and García 2018). It has been shown that Escherichia coli accounts for 30–40% of diarrhoea in developing countries (Odetoyin et al. 2022). An estimate of 60% mortality for non-typhoid Salmonella was reported in African patients with suppressed immune system (Albert et al. 2019).
Generally, β-lactam antibiotics are the frequently used antibacterial agents in treating infections and diseases caused by E. coli and Salmonella species. This has resulted in increased incidence of resistance to these antibiotic classes and therefore, a threat to public health (Serwecinska 2020). Antibiotic resistance in Gram negative bacteria particularly among members of the Enterobacteriaceae family has become a rising problem due to mutation, acquisition of resistance plasmid and other genetic elements encoding resistance genes (Rachel 2018). The clinical use of third generation cephalosporin in the 1980s was noted as a major breakthrough in fighting beta-lactamase producing organism. However, in 1983, plasmid encoding beta-lactamases capable of hydrolyzing cephalosporin was reported (Bush 2018). The most common mechanism of resistance among members of the Enterobacteriaceae is the production of hydrolytic enzymes such as the “β-lactamases” (Ugwu et al. 2020; Bush 2018). Extended spectrum beta-lactamase (ESBL) and Metallo-β-lactamases (MBL) producing organisms emerge due to the production of β-lactamase capable of inactivating the antibiotics used in treatment of infection. Other β-lactam resistance develops through efflux pump, permeability reduction and altered transpeptidases (Bush 2018). Metallo-β-lactamases (MBLs) are β-lactamase enzymes that hydrolyze and confer resistance on carbapenems with the help of zinc ion (Zn2+) as a cofactor for their enzymatic activities (Khalifa et al. 2021).
The emergence and spread of ESBL and MBL-mediated resistances in diarrhoeal causing organisms (e.g. E. coli and Salmonella species) has led to increase in cost of treatment, severity of infection, and duration of diarrhoea episodes (Ugwu et al. 2019; Heredia and García 2018; Odetoyin et al. 2022). Therefore, this study investigated the phenotypic and genotypic characterization of E. coli and Salmonella spp. causing childhood diarrhea in Awka metropolis of Anambra State, Southeastern Nigeria.
Methods
Sampling and ethical approval
The sample size was determined by Cochrane formular using the formular:
where n = sample size, Z = standard normal deviation at 95% confidence interval (which was 1.96), p = proportion of target population, q = alternate proportion (1 − p), e = desired level of precision (degree of precision/significance) = 0.05.
A total of 200 stool samples were used for this present study. This study was done through sampling from three major pediatric tertiary hospitals in Awka metropolis. This sampling was carried out from January, 2018 to April, 2018.
Ethical approval was obtained from the Anambra State Ministry of Health (Ref:MH/AWKA/M:321/131) and was approved by the hospital managements prior to onset of the study. A verbal consent was obtained from the parents/guardians of the under-five subjects and educated on purpose of the research. The information gathered was treated with utmost confidentiality.
Sample collection and isolation
Watery diarrheic Stool samples were collected using a well labeled sterile, transparent, wide mouthed container from children with diarrheic conditions. The samples were transferred in Amies transport media in a sterile zip lock bag and transported to the laboratory under a controlled temperature in a sample box. A loopful of the samples were introduced into a freshly prepared Selenite F broth (Oxoid, UK) and incubated overnight at 37 °C. Isolation of E. coli and Salmonella spp. was based on culture on Eosin methylene blue (EMB) agar, MacConkey agar and Salmonella Shigella agar (Oxoid, UK) according to previous studies (Ugwu et al. 2017; Okoli et al. 2021). Firstly, freshly grown broth cultures were streaked on both agar media using a sterile wire loop. The plates were incubated at 37 °C for 24 h in the incubator (Thermo Fischer Scientific, USA). After incubation, the unique cultural and morphological characteristics of E. coli and Salmonella spp were macroscopically observed on the plates. The suspected colonies were subjected to Gram staining and routine biochemical tests such as indole production, catalase test, oxidase test, and citrate test and hydrogen sulphide production (Ejikeugwu et al. 2014; Tawyabur et al. 2020). All reagents for biochemical tests were procured from Oxoid limited (Oxoid, UK).
Antibiotics susceptibility study of the bacterial isolates
Antibiotics susceptibility profile of all isolates of E. coli and Salmonella were determined by modified Kirby-Bauer disk diffusion method as recommended by the Clinical Laboratory Standard Institute, CLSI (CLSI 2019). The following commercially available antimicrobial disks (Oxoid, UK) were used; ceftazidime (30 µg), cefuroxime (30 µg), gentamicin (10 µg), cefixime (5 µg), ofloxacin (5 µg), amoxicillin-clavulanic acid (30 µg), nitrofurantoin (300 µg), ciprofloxacin (5 µg), imipenem (10 µg), aztreonam (30 µg) and cefpodoxime (10 µg) and metronidazole (50 µg). The isolates were standardized to 0.5 McFarland equivalence and aseptically inoculated on prepared Muller-Hinton agar using a swab stick (Jafari et al. 2020; Ejikeugwu et al. 2014). Single antibiotics disks were aseptically placed on the MH agar plates at a distance of 15 mm apart, and the plated were incubated overnight at 37 °C. The inhibition zone diameters (IZD) were measured, recorded and interpreted based on the standard antibiotic breakpoints of Clinical and Laboratory Standards Institute (CLSI 2019) criteria (CLSI 2019). Escherichia coli ATCC® 25922 and Salmonella enterica subsp. enterica serovar Enteritidis ATCC® 13076 were used as quality control strains for the antimicrobial susceptibility testing (AST) and PCR experiment (Oxoid, UK).
Phenotypic ESBL detection
The isolates which were resistant to 3rd generation cephalosporins were tested for ESBL production using double disks synergy test (DDST) according to the CLSI guidelines. Antibiotic disks of ceftazidime (30 μg) and cefpodoxime (30 μg) were placed on MH agar plate at 16 mm apart center to center from the central amoxicillin-clavulanic acid (20/10 µg) and afterward incubated for 18–24 h at 37 °C. An increase of ≥ 5 mm in the inhibition zone diameter for any of the cephalosporins tested in combination with amoxicillin-clavulanic acid versus its zone when tested singly confirms ESBL production phenotypically (Subedi et al. 2020).
Phenotypic metallo-beta lactamase detection
All isolates that were resistant to imipenem (diameter of zone inhibition ≤ 17 mm) were subjected to imipenem-EDTA combined disc test for MBL production. The test inoculums (0.5 McFarland’s turbidity) were spread onto Muller-Hinton (MH) agar plates by using a sterile cotton swab. Two 10 μg imipenem discs were placed at 20 mm apart on the plate. 5 μl of 0.5 M EDTA (pH 8.0) solution was added to one of the imipenem disc and incubated for 24 h at 37 °C. Enhancement of zone of inhibition of imipenem + EDTA disc compared to that of Imipenem disc alone by ≥ 7 mm was considered MBL positive (Wang and Wang 2020).
Molecular studies
DNA extraction
Fresh overnight culture of bacterial cells was used for this analysis. The DNA of both E. coli and Salmonella were extracted for genetic testing using the Bioneer DNA Genomic Extraction Kit (Bioneer Corp. Korea). The procedures were carried out step by step according to the manufacture’s instruction.
Multiplex PCR amplification
The blaTEM, blaSHV, bla CTX-M and blaVIM genes in E. coli and Salmonella spp were targeted using specific primers shown in Table 1. The PCR master mix was prepared using Accupower@ multiplex PCR Premix (Bioneer, Korea). The Premix tube was added 1 µl of template DNA, 1 µl of each forward and reverse primer, then the tube was completed with nuclease free water. The isolated DNA products were quantified using NanoDrop spectrophotometer (Thermo Fischer Scientific, USA). The final mix for the PCR analysis comprised 26.5 µL of the master mix containing 0.2 µL of Taq polymerase enzyme U/µL, 2.5 µL of 10X PCR buffer along with 2.5 µL MgCl2, 1 µL of 10 pM from each of the forward and reverse primers, 2.5 µL of dNTPs MIX (2 Mm), 3 µL of DNA template (from the test isolates), 14.8 µL of nuclease-free water. A 100 bp DNA molecular marker was used as the positive control while the negative control was a PCR master mix containing distilled water.
Amplification was carried out according to the following thermal cycling condition for Bla gene:
Pre-denaturation of 94 °C for 5 min, followed by 40 denaturation at 94 °C for 30 secs, annealing at 52 °C for 30 secs and 35 cycles of extension for 1 min at 72 °C 35 cycles. The final extension was done at 72 °C for 5 min. Gel electrophoresis of the PCR products was carried out in 1.5% agarose gel for 2 h at 80 V and photographed in a UV transilluminator (Thermo FischerScientific, USA).
Statistical analysis
All statistical analysis were performed using SPSS (version 23) software package and expressed as mean values ± Standard error of the mean of the three replicates of antibiotic susceptibility profile of the isolated bacteria.
Results
Bacteria isolation, characterization and susceptibility testing
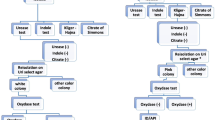
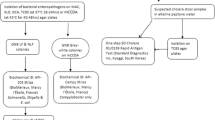
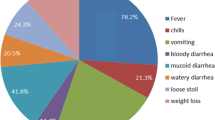
Out of 200 watery diarrheic stool samples collected from children under age five in this study, a total of 157 isolates were identified comprising 49 (24.5%) Salmonella spp and 108 E. coli isolates (54%). The results of the antibiotic susceptibility studies showed that both organisms exhibited remarkable resistance against ceftazidime, cefuroxime, amoxicillin/clavulanic acid and metronidazole. A high resistance of E. coli (95%) to cefuroxime was observed while highest susceptibility was observed in imipenem (88%) followed by nitrofurantoin (84%) and gentamicin (73%). In addition, Salmonella spp was highly resistant (93%) to cefuroxime and were highly susceptible to nitrofurantoin (98%), followed by imipenem (91%), azetronam (85%) and gentamicin (25%) (Figs. 1, 2 and 3).
Phenotypic and genotypic detection of ESBL and MBL
Using double disc synergy test, 45.4% of E. coli and 51.1% of Salmonella spp. were confirmed to be ESBL positive (Fig. 4). Also, 27 (25%) E. coli and 7 (14.3%) Salmonella spp. were identified to be MBL producers (Fig. 5). Similarly, 15 isolates comprising of 11 E. coli and 4 Salmonella spp co-harbored both ESBL and MBL production (Table 2).
The PCR data (Table 3 and Fig. 6) revealed that E. coli harboured the blaVIM 3 (18.8%) and blaSHV 2 (12.5%) genes respectively. Only 1 (6.3%) isolate of E. coli co-harboured bla TEM and bla CTX. The PCR data of ESBL and MBL producing strains of Salmonella spp. revealed that out of 7 isolates studied only 2(28.6%) isolates of the Salmonella species harboured the bla VIM gene while blaSHV gene was discovered in 2(28.6%) isolates of Salmonella species (Fig. 7).
Electrophoretogram showing PCR detection of CTX-M, TEM, VIM and SHV genes in E. coli isolate. Lane M is the DNA marker/ladder. Lane I-8 and 9–15 shows the amplified products CTX-M(550), TEM(918), VIM(437) and SHV(293) genes in E. coli isolates recovered in this study. Lane N is the negative control which contains nuclease free water
Electrophoretogram showing PCR detection of CTX-M, TEM, VIM and SHV genes in Salmonella spp. isolate. Lane M is the DNA marker/ladder. Lane I-7 shows the amplified products CTX-M(550), TEM(918), VIM(437) and SHV(293) genes in Salmonella spp. isolates recovered in this study. Lane N is the negative control which contains nuclease free water
Discussion
One hundred and fifty seven (157) isolates were identified from 200 diarrheic stool samples of which 49 were Salmonella spp. and 108 isolates were E. coli. The occurrence of E. coli was observed to be 54% in the studied population which was higher compared to 36.6% prevalence reported by Adesoji and Liadi (2020) from patients attending Mallam Mande General Hospital in Kastina State, North-Eastern Nigeria and; also 40.7% occurrence was similarly reported by Najim et al., (2019) from healthcare centers in Sokoto. There was no bacterial growth in 53 (26.5%) of the samples which suggested that the cause of the diarrhoea might be due to other causes other than these infectious bacteria. However, 26.5% prevalence was observed in Salmonella spp. which was higher than 8.25% and 5.9% prevalence reported by Pervin et al. (2019) and Najim et al. (2019). The prevalence rate of E. coli (54%) was two times higher than Salmonella spp (24.5%) in our findings. This study has shown that Escherichia coli are the commonest cause of infant diarrhoea in Awka, Nigeria. These inconsistencies of E. coli and Salmonella isolated in different studies may be due to various environmental factors and behavioral pattern of the studied population.
Most isolates of E. coli and Salmonella spp. were highly sensitive to imipenem, nitrofurantoin and gentamicin. In a similar work, a high sensitivity response to these three antibiotics was also reported by Mohammadzadeh et al. (2019). These antimicrobial agents were injectables and prescription only medications; cannot be easily purchased without prescription. These might be one of the few of the reasons why these drugs are highly effective in treating several infections. The diarrheic organisms were also highly resistant to amoxicillin-clavulanic, cefuroxime and ceftazidime. This is similar to the findings of Ugwu et al. (2017) where all these antibiotics showed 100% resistance. It was observed that about 50% E. coli isolates were susceptible to ofloxacin and ciprofloxacin but not for Salmonella that 50% of its isolates were resistant to the same antibiotics. These quinolones are considered a choice of treatment for bacterial infection and yet they are becoming less effective (Guadalupe et al. 2018). Several reports have indicated that these drugs are becoming less effective against other bacteria isolated, largely because of their indiscriminate uses and misuses (Nelson et al. 2019). This increase of resistance is worrisome as we are left with fewer treatment options including the cephalosporins.
In this study, phenotypic analysis showed that the diarrhea infection was caused by 45.4% ESBL producing E. coli and 51.1% ESBL producing Salmonella spp. Lalruatdiki et al. (2018) reported that E. coli had higher percentage of ESBL compared to Salmonella spp. Similarly, Konate et al., (2017) observed a 67% ESBL producing E. coli which is high compared to findings of our study. Difference studies reported that ESBL producer varies from region to region which may be due to difference in local antibiotics prescribing and use patterns (Chander and Shrestha 2013; Bai et al. 2017; Saka et al. 2020). It was observed that ESBL and MBL producers were more resistant to many antibiotics used in this study than non- producers, this is similar to the findings of Ghazaei (2018) that observed a relationship between ESBL production and antibiotics resistance in Salmonella typhi. The prevalence of MBL than ESBL is lower in both organisms with some isolates co-harbouring the β-lactamase genes. In a similar study, Ejikeugwu et al. (2016) reported that 28.6% of E. coli isolated from abattoir was MBL positive.
In this study, we observed in both E. coli and Salmonella that some ESBL studied molecularly harbored at least more than 1 ESBL encoding genes which was similar to the findings of Jafari et al. (2020). The beta-lactamase gene; blaTEM and blaSHV were less common in our settings with less than 50% prevalence. Report from Hamad Medical Corporation, Qatar stated that CTX-M group has evolved through mutations in blaTEM and blaSHV genes and is becoming endemic (Zhou et al. 2018). A study in Pakistan reported a 72% of isolates had blaCTX-M gene (Abrar et al. 2019). In addition, blaVIM, was the β-lactamase gene with the highest prevalence for both E. coli (18.8%) and Salmonella spp. (28.6%). Our reports are similar to the findings of Dembele et al. (2021) in rural area of Burkina Faso. Previously carbapenems resistance Salmonella was rarely isolated but was observed in our study showing a rapid dissemination of these genes (Fernandez 2018). In a related study, the high frequency of multidrug resistant bacteria from urine samples was also reported in a healthcare facility (Bayode et al. 2020). This finding corroborates the high prevalence of multidrug resistant bacteria reported in the current study. The transmission of multidrug resistance genes within the hospital milieus portends grave danger to antimicrobial therapy since it may be difficult to select the best antibiotic for therapy. Babatunde et al. (2022) also used an in silico model to show that genes responsible for multidrug resistance in pathogenic bacteria are spreading fast within the hospital environment. An increase in blaVIM producing organisms is problematic to the clinical settings. It reduces the treatment choices and presents drugs of adverse health effect such as, colistin and polymyxin B as few options (Khosravi and Mihani 2018).
Conclusions
This study showed a high prevalence of diarrhea among children less than 5 years of age in Awka metropolis, South east Nigeria. Escherichia coli were the commonest cause of diarrhoea in Awka, Nigeria, and the species isolated were highly sensitive to imipenem and nitrofurantoin. Both E. coli and Salmonella spp. were found to be multidrug resistant. We therefore advocate for the rational use of available antibiotics in the treatment of diarrhoea-associated infections. The blaVIM was the most prevalent gene while blaCTX-M, blaTEM and blaSHV were less common beta-lactamase genes among the bacterial isolates investigated in this study. Co-expression of multiple antibiotic resistance genes complicates the treatment strategy and requires steady surveillance to reduce its spread.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AST:
-
Antimicrobial susceptibility testing
- ESBL:
-
Extended spectrum beta-lactamase (ESBL)
- MBL:
-
Metallo-β-lactamases
- E. coli :
-
Escherichia coli
- WHO:
-
World Health Organization
- β-lactam:
-
Beta-lactam
- CLSI:
-
Clinical Laboratory Standard Institute
- DDST:
-
Double disks synergy test
- DNA:
-
Deoxyribonucleic acid
- PCR:
-
Polymerase chain reaction
References
Abrar S, Ain NU, Liaqat H, Hussain S, Rasheed F, Riaz S (2019) Distribution of blaCTX− M, blaTEM, blaSHV and blaOXA genes in extended-spectrum-β-lactamase-producing clinical isolates: a three-year multi-center study from Lahore, Pakistan. Antimicrob Res Infect Control 8:80. https://doi.org/10.1186/s13756-019-0536-0
Adesoji AT, Laidi AM (2020) Antibiogram studies of Escherichia coli and Salmonella species isolated fom diarrhoeal patients attending mallam mande general hospital Dusttin-Ma, Kastina State, Nigeria. Pan Afr Med J. https://doi.org/10.11604/pamj.2020.37.11.24851
Albert JM, Bulach D, Alfouzan W, Izumiya H, Carter G, Alobaid K, Alatar F, Sheikh AR, Poirel L (2019) Non-typhoidal Salmonella blood stream infection in Kuwait: clinical and microbiological characteristics. PLoS Negl Trop Dis 13(4):e0007293. https://doi.org/10.1371/journal.pntd.0007293
Babatunde OJ, Okiti AF, Bayode MT et al (2022) Antibiogram profile prediction of selected bacterial strains by in silico determination of acquired antimicrobial resistance genes from their whole-genome sequence. Bull Natl Res Cent 46:230. https://doi.org/10.1186/s42269-022-00922-w
Bai L, Wang L, Yang X, Wang J, Gan X, Wang W, Xu J, Cheng Q, Lan R, Fanning S, Li F (2017) Prevalence and molecular characteristics of extended-spectrum β-lactamase genes in Escherichia coli isolated fom diarrhoeic patients in China. Front Microbiol. https://doi.org/10.3389/fmicb.2017.00144
Bayode M, Olalemi A, Oladejo B (2020) Multiple antibiotic resistant index and detection of QnrS and QnrB genes in bacterial consortium of urine samples from clinical settings. Eur J Biol Res 11:45–56. https://doi.org/10.5281/zenodo.4304311
Bogaerts P, Rezende de Castro R, de Mendonca R, Huang T-D, Glupczynki Y (2013) Validation of carbapemase and extented-spectrum Beta-lactamase multiplex endpoint PCR assay according to ISO 15189. J Antimicrobial Chemother 68(7):1576–1582
Bush K (2018) Past and present perspectives on β- lactamases. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.01076-18
Chander A, Shrestha CD (2013) Prevalence of extended spectrum beta lactamase producing Escherichia coli and Klebsiella pneumoniae urinary isolates in a tertiary care hospital in Kathmandu, Nepal. BMC Res Notes 6(1):487
Clinical and Laboratory Standards Institute (2019) Performance standards for antimicrobial susceptibility testing. In: 29th informational supplement CLSI document M100, Wayne
Dembele R, Soulama I, Aime W, Kabore WA, Konate A, Kagambega A, N’Golo DC, TraoreSeck A, Traore AS, Gussennd N, Gassama-Sow A, Barro N (2021) Molecular characterization of carbepenemase-producing Enterobacterales in children with diarrhoeain rural burkina faso. Drug Deliv Therap 11(1):84–94
Ejikeugwu C, Ugwu M, Iroha I, Eze P, Gugu T, Esimone C (2014) Phenotypic detection of metallo-β-lactamase (MBL) enzyme in Enugu, Southeast Nigeria. Am J Biol Chem Pharm Sci 2(2):1–6
Ejikeugwu C, Edeh C, Iroha I, Orji J, Eluu S, Ugbo E, Duru C, Esimone C (2016) Antibiogram and detection of metallo-beta-lactamase (MBL) positive Escherichia coli isolated from abattoir. Nat Sci 14(11):65–69
Fernandez J, Guerra B, Rodicio MR (2018) Resistance to carbapenems in non-typhodial Salmonella enterica serovars from humans, animals and food. Vert Sci 5:E40. https://doi.org/10.3390/vetsci5020040
Ghazaei C (2018) Phenotypic and molecular detection of β -lactamase genes blaTEM, blaCTX and blaSHV produced by Salmonella spp. isolated from poultry meat. Gene Cell Tissue. https://doi.org/10.5812/gct.84367
Guadalupe PM, Robert MJ, Jorge V, Esvieta T, Fabiola R, Humberto GM, Gilberto Y (2018) Antibiotic susceptibility of Quinolones against Salmonella spp. strains isolated and molecularly sequenced gyrA gene. Microb Pathog 114:286–290
Heredia N, García S (2018) Animals as sources of food-borne pathogens: a review. Anim Nutr 4:250–255
Jafari E, Mostaan S, Bouzari S (2020) Characterization of antimicrobial susceptibility, extended-spectrum β-lactamase genes and phylogenetic groups of enteropathogenic Escherichia coli isolated from patients with diarrhea. Osong Public Health Res Pespect 11(15):327–333
Khalifa SM, AbdEl-Aziz AM, Hassan R, Abdelmegeed ES (2021) β-lactam resistance associated with β-lactamase production and porin alteration in clinical isolates of E. coli and K. pneumonia. PLoS ONE. https://doi.org/10.1371/journal.pone.0251594
Khosravi AD, Mihani F (2018) Detection of metallo-β-lactamase producing Pseudomonas aeruginosa strains isolated from Burn patients in Ahwaz, Iran. Diagnost Microbiol Infect Dis 60(1):125–133
Konate A, Dembele R, Guessennd NK, Kouadio FK, Kouadio IK, Ouattara MB, Kabore WAD, Kagambega A, Cisse H, Ibrahim HA, Bagre TS, Traore AS, Barro N (2017) Epidemiology and antibiotic resistance phenotypesof diarrhoeagenic and Escherichia coli responsible for infantile gastroenterities in Ouagadougou, Burkina Faso. Eur J Microbiol Immun 7(3):168–175
Lalruatdiki A, Dutta TK, Roychoudhury P, Subudhi PK (2018) Extended-spectrum β lactamases producing multidrug resistance Escherichia coli, Salmonella and Klebsiella pneumoniae in pig population of Assam and Meghalaya, India. Vet World 11(6):868–873
Moghaddam MH, Zolfaghari MR, Tavakoli-Hoseini N (2017) CTX-M-15 type β-lactamases from clinical isolates of Escherichia coli by polymerase chain reaction and DNA sequencing. Zahedan J Res Med Sci 19(3):e5814. https://doi.org/10.5812/zjrms.5814
Mohammadzadeh M, Tavakoli M, Yaslianifard S, Asadi E, Golmohammadi R, Mirnejad R (2019) Genetic diversity and antibiotic susceptibility of uropathogenic Escherichia coli isolates from kidney transplant recipients. Infecand Drug Res 12:1795–1803
Najim Z, Kakako LS, Ochei J, Alkali BR, Mohammed K, Opaluwa SA (2019) Prevalence and antibiotic susceptibility pattern of Escherichia coli and Salmonella spp. isolated from diarrhoeic children in selected health centers in Sokoto, Nigeria. Asian J Res Infect Dis 2(1):1–9
Nelson DW, Moore JE, Rao JR (2019) Antimicrobial resistance (AMR): significance to food quality and safety. Antimicrobiol Res 3(1):17–22
Odetoyin B, Ogundipe O, Onanuga A (2022) Prevalence, diversity of diarrhoeagenic Escherichia coli and associated risk fastors in well water in Ile-Ife, Southern Nigeria. One Heath Outlook 4:3. https://doi.org/10.1186/s42522-021-00057-4
Okoli MF, Ugwu MC, Ezejiegu KC et al (2021) Antibiotic susceptibility profile of Salmonella spp and Shigella spp isolated from commercial frozen chicken sold in three markets within Awka Metropolis. Clin Immunol Res 5(2):1–4
Paykoc EL, Turkyilmaz S (2018) Investigation of P fimbriae presence in Escherichia coli strains isolated from urine samples in human and their antibacterial resistance. Jundishapur J Microbiol 11(9):e66119
Pervin MK, Jhora ST, Paul S, Naher A (2019) Antibiotic susceptibility pattern of diarrhoeal pathogens in under five children. Bang Med J Khulna 52:35–39
Rachel L, Medernach MD, Latania K, Logan MD (2018) The growing threat of antibiotic resistance in children. Infect Dis Clin N Am 32(1):1–17. https://doi.org/10.1016/j.idc.2017.11.001
Saka HK, Gracia-Soto S, Dabo NT, Lopez-Chavarrias V, Muhammed B, Ugarte-Ruiz M, Alvarev J (2020) Molecular detection of extended spectrum β-lactamase genes in Escherichia coli isolates from diarrhoeic children in Kano, Nigeria. PLoS ONE. https://doi.org/10.1371/journal.pone.0243130
Serwecinska L (2020) Antibacterial and antibiotic-resistant bacteria: a risk to the environment and to public health. Water 12:2213. https://doi.org/10.3390/w12123313
Subedi K, Karki F, Lama S, Pandey A, Dahal U, Paudyal R (2020) Phenotypic detection of extended spectrum beta-lactamase production from E. coli in urinary sample among children. Tribhuvan Univ J Microbiol 7(1):75–83
Tawyabur M, Islam S, Sobur A, Hossain J, Mahmud M, Paul S, Hossain T, Ashour M, Rahman T (2020) Isolation and characterization of multidrug-resistant Escherichia coli and Salmonella spp. from healthy and diseased turkeys. Antibiotics 9:770–784
Tsehay CT, Aschalew AY, Dellie E, Gebremedhin T (2021) Feeding practices and associated factors during diarrhoeal disease among children aged less than five years: evidence from the Ethopian Demographic and Health Survey 2016. Ped Health Med Thera 12:69–78
Ugboko HU, Nwinyi OC, Oranusi SU, Oyewale JO (2020) Corrigendum to “childhood diarrhoeal diseases in developing countries.” Heliyon 6(4):e03690
Ugwu MC, Edeani GI, Ejikeugwu CP, Okezies U, Ejiofor SO (2017) Antibiotic susceptibility profile of Escherichia coli and Salmonella causing childhood diarrhoea in Awka municipal, South-eastern Nigeria. Clin Microbiol 6:277. https://doi.org/10.4172/2327-5073.1000277
Ugwu MC, Omanukwue C, Chimezie C, Okezie U, Ejikeugwu CP, Nnabuife-Illoh E, Esimone CO (2019) Poultry farm and poultry products as sources of multiple antimicrobial-resistant Salmonella and S. aureus. J Trop Dis 7:308. https://doi.org/10.4172/2329-891X.1000308
Ugwu MC, Shariff M, Nnajide CM, Beri K, Okezie UM, Iroha IR, Esimone CO (2020) Phenotypic and molecular characterization of β-lactamases among enterobacterial uropathogens in South-eastern Nigeria. Can J Infect Dis Med Microbiol. https://doi.org/10.1155/2020/5843904
Ugwu MC, Ofoegbu E, Ezejiegu KC (2020) Spectrum and antibiogram of bacteria isolated from commercially available stockfish in Eke-Awka Market, Anambra Nigeria. Acta Sci Microbiol 3(10):22–26
Wang W, Wang X (2020) Prevalence of metallo-betalactamase genes among Pseudomonas aeruginosa Isolated from various cliniical sample in China. J Lab Med 44(4):197–203
World Health Organization and Child Epidemiology Estimation Group (MCEE). Estimate (2018). Estimate of child cause of death, Diarrhoea 2018. http://apps.who.int/gho/data/node.main.ChildMort?lang=en. Accessed 23 Aug 2018
Zhou Y, Zhu X, Hou H, Lu Y, Yu J, Mao L, Mao L, Sun Z (2018) Characteristics of Diarrhoeagenic Escherichia coli among children under 5years of age with acute Diarrhoea: a hospital based study. BMC Infect Dis 18:63–73
Acknowledgements
The authors are grateful to Anambra State Ministry of Health, Awka (Ref:MH/Awka/M:321/131) and the hospital managements of Chukwuemeka Odumegwu Ojukwu University Teaching Hospital, Awka (COOUTH), Prime Specialist Hospital, Awka and Covenant Hospital Okpuno, Awka
Funding
Not applicable/No funding was received.
Author information
Authors and Affiliations
Contributions
IEN: performed sample collection, microbial identification and antibiotic susctibility Data curation. He also participated in writing- Original draft preparation. MCU: Developed the Concept and was involved in Writing- Reviewing and Editing the manuscript. PCE: was a major contributor in writing Reviewing and Editing the manuscript. NTU: participated in Data curation. IRI: Supervised the work and reviewed the manuscript. COE: was a key contributor in Conceptualization & Supervision of the work. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical approval was obtained from the Anambra State Ministry of Health (Ref:MH/Awka/M:321/131) and was approved by the hospital managements prior to onset of the study. A verbal consent was obtained from the parents/guardians of the under-five subjects about the purpose of the research.
Consent for publication
Not applicable.
Competing interests
No competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nwike, I.E., Ugwu, M.C., Ejikeugwu, P.C. et al. Phenotypic and molecular characterization of enteropathogenic Escherichia coli and Salmonella spp. causing childhood diarrhoea in Awka, South-Eastern Nigeria. Bull Natl Res Cent 47, 97 (2023). https://doi.org/10.1186/s42269-023-01076-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42269-023-01076-z