Abstract
Background
Numerous studies have investigated the role of apolipoprotein E (APOE) polymorphisms in coronary artery disease (CAD), but some controversies exist regarding the outcomes as the results were not consistent and remain uncertain. Therefore, the present meta-analysis was conducted to evaluate the association of APOE polymorphisms with coronary artery disease.
Methods
All the relevant studies published in English language till August 2020 were identified by searching through various electronic databases. The complete data was independently extracted by the two researchers. The data were analyzed by using the Comprehensive Meta-Analysis program and MetaGenyo program. The pooled odds ratio was used to check the associations between CAD and APOE polymorphisms. The following genetic models were used to calculate the odds ratio: ε2 vs. ε3 and ε4 vs. ε3.
Results
In the final analysis, we include 12 studies regarding the role of APOE polymorphism in CAD. The pooled odds ratio for ε4 allele was higher (OR 2.00; 95% and CI, 1.48–2.71). There is no statistical significant association for ε2 allele with CAD (OR 1.38; 95% CI, 1.18–1.62). This analysis showed no publication bias exists in the current meta-analysis.
Conclusions
Our findings suggest that the apolipoprotein ε4 allele appears as a significant genetic risk factor for coronary artery disease while the ε2 allele is beneficial to alleviate the CAD risk.
Trial registration
Registered with PROSPERO International Prospective Register of Systematic Reviews. PROSPERO registration number CRD42020190464
Similar content being viewed by others
Background
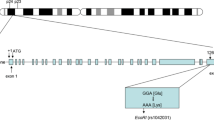
The cardiovascular system is comprised of vessels and heart, which lead the foundation of cardiovascular diseases (CVDs) grouping into the two systems, i.e., circulatory and heart system. In 1772, angina clinical description was provided by Heberden and almost over a century, pathologists gave attention to the coronary artery blockade events [1]. Coronary artery disease (CAD) is the most common form of cardiovascular disorder and a leading cause of mortality occurring worldwide. Its prevalence rate is ranked highest in developing countries as it majorly affects the adult human population [2]. The World Health Organization (WHO) positioned CAD on number three in a list of the most prevailing disorders. In relation to non-communicable diseases (NCDs), CAD alone contributes 19% of the total economic burden. The mortality rate is seen highest in South Asian countries and lowest in the east [3]. CAD is identified as a heritable multifactorial disorder that instigated via a complex interaction between genetic and environmental factors [4]. A lot of evidence based information is already available on the pathogenesis of CAD and its risk factors, i.e., gender, age, hypertension, serum cholesterol levels, and smoking. Among all the risk factors, atherosclerosis is being considered as a key etiological factor that contributes in the initiation of CAD [5]. Further, disparities in ethnic groups also have an influence on the development and progression of CAD. Differences in ethnic races resulted in the greater severity and prevalence in certain human communities of both developed and developing countries. These ethnic races rather have differences in their cultures, customs, lifestyles and eating habits which at different levels offer exposure to CAD risk factors [6]. Recently, by using the genome wide association studies several attempts have been made to untwist CAD association with the several genetic loci involved in the pathogenesis of atherosclerosis. Researchers are concentrating more on extensive CAD genetics as data obtained from such findings would be helpful in the future for the prevention and treatment of the disease [7]. In addition, high lipid levels in the blood are also main variable risk factors that could instigate CAD. The low-density lipoprotein cholesterol (LDL-C) is an independent risk factor and also a novel biomarker for CAD. The drugs that lower the serum LDL-C are beneficial in the reduction of CAD risk [8]. Single-nucleotide polymorphism (SNPs) can be used as a tool to evaluate the possibility of developing CAD in relation to the genetic biomarkers. Many SNPs are located on the genes which are inherited independently and are involved in regulation of the plasma lipid concentration [9]. Apolipoprotein E (APOE) is 299 amino acids containing apolipoprotein which is small in size and synthesized in the liver. It is an important constituent of various lipoproteins, including very low-density lipoprotein (VLDL), high-density lipoprotein (HDL), and chylomicrons. APOE plays an essential role in the metabolism of LDL by enhancing its cellular uptake. In inflamed arteries, APOE is also produced by locally residing macrophages, which facilitates in removing of atherosclerosis clots by lowering the cholesterol levels in blood [10]. The APOE gene is located on chromosome 19, APOE polymorphisms (rs7412 and rs429385) resulted into the altered amino acids arrangement at position 130 and 176 as a consequence three different protein isoforms are produced which are called as APOE2, APOE3, and APOE4.
Rationale
Many findings have confirmed that APOE4 (rs429385) causes an increase in concentration of LDL-C while APOE2 (rs7412) decreases LDL-C concentration. The isoform ε3 is the wild type which modulates the clearance of lipoproteins from plasma as it has normal affinity for low-density lipoprotein receptors. When the 158th amino acid is replaced by cysteine ε3 mutates into ε2 and when 112th amino acid is replaced by arginine it mutates into ε4. Moreover, distribution of these alleles is varied among different ethnic populations, i.e., Asians have low incidence of ε2 and ε4 as compared to Africans and Europeans [11,12,13]. Since, the meta-analysis on genetic association of polymorphism in APOE gene with CAD is scarce. Thus, it is the need of the hour to analyze the available literature. We designed the systematic review and meta-analysis to check the significant association of polymorphisms in APOE gene with CAD that provides us the management, prevention, and better insight into the role of genetic risk factors in causing the CAD.
Objectives
The objective of the present study was to evaluate the association of genetic polymorphisms in APOE gene with coronary artery disease.
Methods
In the current study, we followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses 2009 (PRISMA 2009) statement. A PRISMA checklist is provided as supplementary file.
Search strategy
We searched all published articles in MEDLINE, EMBASE, PubMed, Ovid Web of Science, and the Cochrane Library (up to 14 august 2020) using the following MeSH terms and keywords: including ‘coronary artery disease,’ ‘CAD,’ ‘APOE,’ ‘Apolipoprotein E,’ ‘gene polymorphism,’ ‘variant,’ ‘genotype,’ ‘mutation,’ ‘atherosclerosis,’ ‘coronary heart disease,’ and ‘worldwide.’ In addition, to avoid missing any relevant study, we have searched the related articles manually. All the duplicate studies were not included in the final analysis.
Inclusion and exclusion criteria
The extracted articles were included in this study when the following criteria were met: (1) retrospective case-control studies using either a population-based design or hospital-based design. (2) The full length original articles on the association of APOE polymorphisms and CAD in human subjects. (3) Sufficient information was provided for estimating the statistical analysis including odds ratio (OR). Those provided no sufficient data or not written in English language and designed as systematic reviews, meta-analysis, or not designed as case-control studies were eliminated.
Data extraction
To reduce the selection bias, the authors independently reviewed and extracted the data required for all the included studies using a predesigned data extraction table. From each original study, the following data were abstracted: author names, year of publication, country and ethnicity, baseline characteristics, sample size, method, the distribution of genotypes and alleles in subjects, and evidence of conforming to the Hardy-Weinberg equilibrium (HWE). Moreover, in the included studies the criteria for the diagnosis of the CAD was ≥ 50% angiographically documented luminal stenosis in at least one of the three major coronary arteries. The detailed characteristics of included study are given in Tables 1 and 2.
Quality score assessment
We used the Newcastle-Ottawa Scale (NOS) to determine the quality of each included study. The NOS ranges between 0 (worst) and 9 stars (best). The quality of the articles was independently assessed by the authors and any discrepancies were decided through discussion to achieve a consensus.
Statistical analysis
The association of the APOE polymorphism and risk of CAD was estimated by calculating the pooled ORs and 95%CI. The heterogeneity among the included studies was calculated using the chi-square test and I2 statistic. A fixed-effect model (Mantel–Haenszel) was used in the absence of heterogeneity and if heterogeneity is present, then the random effect model (DerSimonian–Laird) was adopted to investigate the variation both from in-study and between-study. This method was used for various genetic models and for separate analysis of 휀2 and 휀4 carrier status. Furthermore, to assess the stability of results the sensitivity analysis was performed. Publication bias was analyzed by using the funnel plot which was calculated by using Begg’s and Egger’s tests. All statistical analysis was performed by using the Comprehensive Meta-Analysis program (Biosta, Englewood, NJ, USA) and MetaGenyo program.
Results
Characteristics of studies
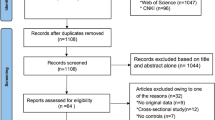
Based on search strategy, 377 potentially eligible studies were included in the initial search. After a comprehensive literature search applying our inclusion criteria, 12 relevant articles were enrolled in final systematic review and meta-analysis. The study selection process is described in Fig. 1. The characteristics of the included studies [13,14,15,16,17,18,19,20,21,22,23,24], and the results of quality assessment based on NOS score are given in Table 1. The frequencies of APOE allele and distribution of genotypes are given in Table 2.
Association between ε2 allele and CAD
The results of association between ε2 allele and coronary artery disease are shown in Fig. 2. We chose fixed-effects models to merge all data based. Overall, the ε2 allele and its carriers had a protective effect on CAD (OR 1.38; 95% CI, 1.18–1.62). The I2 value was 43.1%, tau-squared was 0.0633 and the p value was 0.0554.
Association between ε4 allele and CAD
The results of association between ε4 allele and coronary artery disease are shown in Fig. 3. To merge the entire data, we adopted the fixed effects models. Overall, the ε4 allele and its carriers have a statistical significant effect on CAD (OR 2.00; 95% and CI, 1.48–2.71). There was no heterogeneity observed as the I2 value was 26.2%, tau-squared was 0.106 and the p value was 0.1869.
Sensitivity analysis
Sensitivity analyses showed that after excluding each literature the overall calculated OR did not change significantly, which confirmed the reliability and stability of our analysis.
Publication bias
The shapes of funnel plots do not show any apparent asymmetry in all the genetic models, furthermore, the statistical results also confirmed the absence of publication bias as p > 0.05 for both Begg and Egger tests (Figs. 4 and 5).
Discussion
CAD is a polygenetic and heterogeneous disorder that is triggered as a result of gene-environment and gene-gene multifarious interactions. Since, several investigations have demonstrated an association between CAD incidence and APOE gene polymorphisms, still a lot of controversies exist between the results of these studies. Furthermore, reliability of findings is remained irresolute which are obtained from the single case-referent investigations due to limited population sample size. An increased sample size of population would be helpful to solve this issue and make the data more reliable. Added into it, meta-analysis of larger population sample size has ability to pinpoint even the minor effects of studies that were conducted to evaluate the relationship of diseases to human genetics. The purpose of the current study is to determine the association of APOE gene polymorphisms with coronary artery disease.
To the best of our knowledge, this present meta-analysis provides the most comprehensive assessment of CAD association with SNPs in APOE gene. Our study results revealed that the risk of developing coronary artery disease in ε4 allele carriers was higher as compared to the ε2 allele. We found no association between APOE polymorphism and any of the traditional cardio-vascular risk factors including BMI. Many studies have examined APOE with lipid metabolism, particularly low density lipoprotein cholesterol levels, and CAD risk. Still, there is a lack of information by which ε4 allele carriers modulate the adverse lipid profiles [25]. The APOE polymorphisms are also associated with other pathological conditions, including Alzheimer’s disease, schizophrenia, Parkinson’s disease, diabetes, and beta-thalassemia [13].
The human apolipoprotein E is a serum glycoprotein which has many biological properties. It is approximately of 35 kDa glycosylated protein and the normal function of all the 3 isoforms of a gene is related to their receptor affinity [26, 27]. The ε4 allele is seen as a ‘thrifty’ gene and the key functions of this allele are to raise the production of cholesterol in the liver [27, 28] also reported that it has a major function on lipid clearance and metabolism in humans. The result of their study is also in accordance to our study. They reported that ε2 allele is not associated with CAD as it decreases the LDL levels in plasma. On the other side, ε4 is a risk factor for high levels of LDL cholesterol, total cholesterol, and very low-density lipoprotein cholesterol levels in plasma as it suppresses the synthesis of LDL-receptors (LDLRs) [27].
Similarly, the present study results are in accordance with the previous studies, including meta-analysis performed by Wang et al. report that ε2 allele is a protective factor, while ε4 allele is a risk factor of CAD, especially in Caucasian and Asian population [29]. Additionally, our results are also in accordance with those obtained by Song et al. [30]. Zahng et al. also showed that the allele ε2 has no significant association; however, ε4 allele is a main risk factor for CAD in the Chinese population [31]. Similarly a meta-analysis was conducted by the Wilson to assess the impact of apolipoprotein E alleles on coronary artery disease and the findings of this study also report that ε4 allele was associated with coronary artery disease [32]. It is important to evaluate the publication bias that may introduce false positive results in a meta-analysis [33]. So, there is a need for statistical analysis to avoid possible bias in publications. In order to avoid this bias, we performed Begg and Egger test for investigating the bias in selected studies and we found no obvious publication bias.
Limitations
The results of the current meta-analysis are quite inclusive; however, certain limitations are also obvious in this study. First, in the current study, we mainly focused on APOE gene SNPs and were not able to check other genes responsible for coronary artery disease. Second, we only choose articles that were published in English language and possibly this can result the publication biasness.
Conclusion
It is concluded that the ε4 allele is a chief reason to instigate coronary artery disease while the ε2 allele is beneficial to mitigate the CAD risk. Further, it is suggested that there is a need for more studies with larger sample size in different subgroups with various covariates, which could be helpful to get a more definite conclusion.
Availability of data and materials
The data and materials will be available with the corresponding author upon reasonable request.
Abbreviations
- APOE :
-
Apolipoprotein E
- CAD:
-
Coronary artery disease
- CVDs:
-
Cardiovascular diseases
- WHO:
-
World Health Organization
- NCDs:
-
Non-communicable diseases
- LDL-C:
-
Low-density lipoprotein cholesterol
- SNPs:
-
Single-nucleotide polymorphisms
- VLDL:
-
Very low-density lipoprotein
- HDL:
-
High-density lipoprotein
- PRISMA 2009:
-
Preferred Reporting Items for Systematic Reviews and Meta-Analyses 2009
- OR:
-
Odds ratio
- HWE:
-
Hardy-Weinberg equilibrium
- NOS:
-
Newcastle-Ottawa scale
References
Nabel EG, Braunwald E (2012) A tale of coronary artery disease and myocardial infarction. N Engl J Med. 366:54–63. https://doi.org/10.1056/NEJMra1112570
Shabana N, Ashiq S, Ijaz A, Khalid F, Ul Saadat I, Khan K, et al. Genetic risk score (GRS) constructed from polymorphisms in the PON1, IL-6, ITGB3, and ALDH2 genes is associated with the risk of coronary artery disease in Pakistani subjects. Lipids Health Dis. 2018;17:224. https://doi.org/10.1186/s12944-018-0874-6
Ashiq S, Ashiq K, Shabana S, Shahid SU, Qayyum M, Sadia H (2020) Prevalence and role of different risk factors with emphasis on genetics in development of pathophysiology of coronary artery disease (CAD). Pak Heart J. 52:279–287
Ozaki K, Tanaka T. Molecular genetics of coronary artery disease. J Hum Genet. 2016; 61(1):71-77. https://doi.org/10.1038/jhg.2015.70
Liu Z, Pericak-Vance MA, Goldschmidt-Clermont P, Seo D, Wang L, Rundek T, et al. Coronary collateralization shows sex and racial-ethnic differences in obstructive artery disease patients. PLoS One. 2017;12:e0183836. https://doi.org/10.1371/journal.pone.0183836
Abbasi SH, Sundin Ö, Jalali A, Soares J, Macassa G. Ethnic differences in the risk factors and severity of coronary artery disease: a patient-based study in Iran. J Racial Ethn Health Disparities. 2018;5:623-631. https://doi.org/10.1007/s40615-017-0408-3
Khera AV, Kathiresan S. Genetics of coronary artery disease: discovery, biology and clinical translation. Nat Rev Genet. 2017;18:331-344. https://doi.org/10.1038/nrg.2016.160
Field PA, Vasan RS. LDL-cholesterol is not the only clinically relevant biomarker for coronary artery disease or acute coronary syndrome. Clin Pharmacol Ther. 2018;104:232-234. https://doi.org/10.1002/cpt.1126
Shahid SU, Cooper JA, Beaney KE, Li K, Rehman A, Humphries SE. Effect of SORT1, APOB and APOE polymorphisms on LDL-C and coronary heart disease in Pakistani subjects and their comparison with Northwick Park Heart Study II. Lipids Health Dis. 2016;15:83. https://doi.org/10.1186/s12944-016-0253-0
Pleva L, Kusnierova P, Plevova P, Hilscherova S, Karpisek M, Zapletalova J, et al. The APOE ɛ2 allele is associated with increased plasma apolipoprotein E levels in patients with coronary artery disease. Cor et Vasa. 2017;59:e235-e2e9. https://doi.org/10.1016/j.crvasa.2016.11.011
Burman D, Mente A, Hegele RA, Islam S, Yusuf S, Anand SS. Relationship of the ApoE polymorphism to plasma lipid traits among South Asians, Chinese, and Europeans living in Canada. Atherosclerosis. 2009;203:192-200. https://doi.org/10.1016/j.atherosclerosis.2008.06.007
Eichner JE, Dunn ST, Perveen G, Thompson DM, Stewart KE, Stroehla BC. Apolipoprotein E polymorphism and cardiovascular disease: a HuGE review. Am Journal Epidemiol. 2002;155:487-495. https://doi.org/10.1093/aje/155.6.487
Larifla L, Armand C, Bangou J, Blanchet-Deverly A, Numeric P, Fonteau C, et al. Association of APOE gene polymorphism with lipid profile and coronary artery disease in Afro-Caribbeans. PLoS One. 2017;12:e0181620. https://doi.org/10.1371/journal.pone.0181620
Abd El-Aziz TA, Mohamed RH. LDLR, ApoB and ApoE genes polymorphisms and classical risk factors in premature coronary artery disease. Gene. 2016;590:263-269. https://doi.org/10.1016/j.gene.2016.05.032
Afroze D, Yousuf A, Tramboo NA, Shah ZA, Ahmad A. ApoE gene polymorphism and its relationship with coronary artery disease in ethnic Kashmiri population. Clin Exp Med. 2016; 551-556. https://doi.org/10.1007/s10238-015-0389-7
Atis O, Sahin S, Ceyhan K, Ozyurt H, Akbas A, Benli I (2016) The distribution of apolipoprotein E gene polymorphism and apolipoprotein E levels among coronary artery patients compared to controls. Eurasian J Med. 48(2):90–94. https://doi.org/10.5152/eurasianjmed.2015.25
Cheema AN, Bhatti A, Wang X, Ali J, Bamne MN, Demirci FY, et al. APOE gene polymorphism and risk of coronary stenosis in Pakistani population. Biomed Res Int. 2015. https://doi.org/10.1155/2015/587465
Long Y, Zhao XT, Liu C, Sun YY, Ma YT, Liu XY, et al. A Case-Control Study of the Association of the Polymorphisms of MTHFR and APOE with Risk Factors and the Severity of Coronary Artery Disease. Cardiology. 2019;142:149-157. https://doi.org/10.1159/000499866
Ghaznavi H, Kiani AA, Soltanpour MS. Association study between DNA methylation and genetic variation of APOE gene with the risk of coronary artery disease. Mol Biol Res Commun. 2018;7:173-179. doi: https://doi.org/10.22099/mbrc.2018.30955.1352
Arafa S, Abdelsalam S, El-Gilany AH, Mosaad YM, Abdel-Ghaffar A. Endothelial nitric oxide synthase Glu 298 Asp (G894T) and Apolipoprotein E gene polymorphism as possible risk factors for coronary heart disease among Egyptians. Egypt Heart J. 2018;70:393-401. https://doi.org/10.1016/j.ehj.2018.08.001
Heidari MM, Foruzannia SK, Khatami M, Hadadzadeh M, Meybodi ME (2013) Apolipoprotein E gene polymorphism in Iranian coronary atherosclerosis patients candidate for coronary artery bypass graft. Iran J Basic Med Sci. 16:841–844
Hou J, Deng Q, Guo X, Deng X, Zhong W, Zhong Z. Association between apolipoprotein E gene polymorphism and the risk of coronary artery disease in Hakka postmenopausal women in southern China. Lipids Health Dis. 2020;19:1-7. https://doi.org/10.1186/s12944-020-01323-6
Souza DR, Nakachima L, Biagioni RB, Nakazone MA, Pinhel MA, Trindade DM, et al. Relevance of apolipoprotein E4 for the lipid profile of Brazilian patients with coronary artery disease. Braz J Med Biol Res. 2007;40:189-197. https://doi.org/10.1590/S0100-879X2006005000065
Đjan I, Stokić E, Sakač D, Đan M, Obreht D, Erak M, et al. Case-control study of apoE gene polymorphism in young CHD patients and controls in the Serbian population. Arch Biol Sci. 2011;63:89-98. https://doi.org/10.2298/ABS1101089D
Anoop S, Misra A, Meena K, Luthra K (2010) Apolipoprotein E polymorphism in cerebrovascular & coronary heart diseases. Indian J Med Res. 132:363–378
Chaudhary R, Likidlilid A, Peerapatdit T, Tresukosol D, Srisuma S, Ratanamaneechat S, et al. Apolipoprotein E gene polymorphism: effects on plasma lipids and risk of type 2 diabetes and coronary artery disease. Cardiovasc Diabetol. 2012;11:36. https://doi.org/10.1186/1475-2840-11-36
Utermann G, Hees M, Steinmetz A. Polymorphism of apolipoprotein E and occurrence of dysbetalipoproteinaemia in man. Nature. 1977;269:604-607. https://doi.org/10.1038/269604a0
Mahley RW, Rall Jr SC. Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genom Hum G. 2000;1:507-537. https://doi.org/10.1146/annurev..genom.1.1.
Wang YL, Sun LM, Zhang L, Xu HT, Dong Z, Wang LQ, et al. Association between Apolipoprotein E polymorphism and myocardial infarction risk: A systematic review and meta-analysis. FEBS Open Bio. 2015;5:852-858. https://doi.org/10.1016/j.fob.2015.10.006
Song Y, Stampfer MJ, Liu S. Meta-analysis: apolipoprotein E genotypes and risk for coronary heart disease. Ann Intern Med. 2004;141:137-147. https://doi.org/10.7326/0003-4819-141-2-200407200-00013
Zhang MD, Gu W, Qiao SB, Zhu EJ, Zhao QM, Lv SZ. Apolipoprotein E gene polymorphism and risk for coronary heart disease in the Chinese population: a meta-analysis of 61 studies including 6634 cases and 6393 controls. PloS one. 2014;9:e95463. https://doi.org/10.1371/journal.pone.0095463
Wilson PW, Schaefer EJ, Larson MG, Ordovas JM. Apolipoprotein E alleles and risk of coronary disease: a meta-analysis. Arterioscler Thromb Vasc Biol. 1996;16:1250-1255. https://doi.org/10.1161/01.ATV.16.10.1250
Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629-34. doi: https://doi.org/10.1136/bmj.315.7109.629
Acknowledgements
Not applicable
Funding
Not applicable
Author information
Authors and Affiliations
Contributions
SA designed the study. SA and KA retrieved the articles, extracted the data from the included studies, wrote, and reviewed the final manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
PRISMA 2009 Checklist
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ashiq, S., Ashiq, K. The association of apolipoprotein-E (APOE) gene polymorphisms with coronary artery disease: a systematic review and meta-analysis. Egypt J Med Hum Genet 22, 16 (2021). https://doi.org/10.1186/s43042-021-00135-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-021-00135-2