Abstract
Background
Sensorineural hearing loss (SNHL) is a clinically and genetically heterogeneous group of disorders of the auditory system. SNHL can occur as a symptom in more than 400 syndromes, and mutations in more than 150 genes can lead to SNHL. Mutations in the GJB2 and GJB6 genes are among the most common causes of SNHL worldwide. Mutations in Cadherin 23 (CDH23) can cause Usher syndrome and/or non-syndromic hearing loss (NSHL).
Material and methods
In this study, the Whole Exome Sequencing (WES) was used to detect the cause of hearing loss in a large consanguineous Iranian family with two patients. All family members underwent a thorough Genotype–phenotype correlation assessment and co-segregation analysis to understand the inheritance pattern within the family. The candidate variants were further confirmed by Sanger sequencing. In addition, in silico analysis was performed to predict the functional impact of the variants; the interpretation of the variants was performed in accordance with the American College of Medical Genetics (ACMG) guidelines.
Results
WES results identified two novel variants, a homozygous missense variant in CDH23 (c.2961T > G) and a heterozygous splice site variant in OTOGL that was compatible with the autosomal recessive pattern of inheritance. Bioinformatics studies confirmed the pathogenic effects of novel variants. The c.2961T > G variant was classified as likely pathogenic.
Conclusions
The novel identified variant in the CDH23 was the cause of congenital profound progressive form of HL. Samples were not available from the second family to distinguish which variant is responsible for the molecular pathology of the disease. Further studies and functional examinations are suggested for investigating the role of OTOGL: c. 1863-1G > T in deafness.
Similar content being viewed by others
Introduction
Sensorineural hearing loss (SNHL) is the most common sensory disorder in humans [1]. Genetic deficits, structural anomalies, some viral infections, and teratogens account for the etiology of Congenital hearing loss (HL) [2,3,4]. Approximately 1 to 2 out of every 1000 live births are affected by congenital HL. About 67% O HLs etiology attributed to genetic defects [5]. HL is classified into two types: syndromic, 30% of the cases in which, HL occurs as a feature of a syndrome, in more than 400 syndromes, and non-syndromic which contains 70 of the remaining cases [6].
To date, mutations in more than 150 genes have been identified to cause non-syndromic HL [7]. Inheritance modes of SNHL include: autosomal dominant (DFNA, about 17%), autosomal recessive (DFNB, about 80%), X-linked (DFNX, about 1–2%), and mitochondrial (about 1%). Therefore, the autosomal recessive mode of inheritance is the most common cause of the HL among studied cases [8]. Autosomal recessive SNHL (ARSNHL) can occur as a result of homozygous and compound or double heterozygous mutations in some genes [9]. Mutations in GJB2 and GJB6 gene have been identified as the prevalent causes of SNHL in many studied populations [10,11,12]. To date, there is no formal report of GJB6 mutations from Iran, while GJB2 mutations are frequently reported from different parts of Iran among different ethnics and races [13, 14].
At the molecular level, SNHL is the result of defects in genes with diverse functions such as those encoding: transcription factors, POU3F4 and ESRRB [15], gap junction proteins GJB2 and GJB6), motor proteins, MYO6A, MYO7A [16], cell–cell adhesion molecules, CHD23, anion transporters, integral membrane proteins, TMIE, TMC1, extracellular matrix proteins TECTA, OTOA [17, 18], signaling molecules, PJVK[19], methyl transferase LRTOMT [20], serine protease, TMPRSS3, and cytoskeleton proteins such as TRIOBP.
CDH23 is an atypical member of cadherin superfamily; its encoding gene (CHD23 (NM_022124) located on 10q22.1 and includes 70 exons. CDH23 is a calcium dependent cell–cell adherent glycoprotein with a long extracellular tail. It plays essential roles in stereocilia organization and inner ear function. Mutations in CDH23 can affect mechano-transduction and tip links of sensory hair cells in mice [21,22,23].
OTOGL, which is located on human chromosome long arm 12 (12q.21.31( encodes a protein containing 2344 amino acids known as Otogelin-like protein (OTOGL) [24, 25]. OTOGL is a component of tectorial membrane that is expressed in the inner ear of vertebrates in high levels in embryonic stages. OTOGL and Stereocilin interactions are essential in the development of outer hair cells of the cochlea, vestibular system, and inner ear cells, mutations in which would lead to congenital mild-to-moderate hearing loss. Knockdown of OTOGL in zebra fish has been reported to result in moderate SNHL [26,27,28,29]. In human, mutations in different parts of the OTOGL have been reported as the cause of mild-to-moderate non-progressive autosomal recessive HL DFNB84B [30] and retinal disease [31].
The next-generation sequencing (NGS) is a high-throughput technology, that helps the researchers discover the genetic etiology of human diseases and genome variants, especially in the case of complex and high heterogeneous disorders such as HL [32, 33] and intellectual disability [34]. Due to high locus heterogeneity of HLs, whole exome sequencing (WES) as the most common approach of NGS was carried out to define the genetic etiology of a profound congenital form of HL in one single extended Iranian family.
Material and methods
Subjects, DNA extraction, GJB2 analysis
This study was approved by the Ethics Committee of the Isfahan University of Medical Sciences (IR.MUI.RESEARCH.REC.1397.109).
Two patients suffering from two forms of ARNSHL including congenital profound (IV3) and moderate (III3) HL and normal members of a large inbreed Iranian family with multi consanguineous marriages from Isfahan, center of Iran, were recruited to examine for HL and retinopathy and evaluation of thyroid function. Genetic counseling was done and family information was obtained, pedigree was drawn by our genetic counselor (Fig. 1).
After taking informed consent, 5 ml peripheral blood sample was taken from all the consent members of the family and conserved in EDTA -continuing tubes. DNA was extracted from peripheral blood samples by a DNA extraction kit (BioGenet Korea). Purity and concentration of DNA samples were determined using 1% agarose gel and Nanospec cube biophotometer (Nanodrop 2000 Thermoscientific, USA), respectively. The GJB2 gene was amplified using our previous studied primers [35]. Sanger sequencing was done to find the probable mutations in the GJB2 gene by comparing the sequences results with the genomic reference sequence, NG_008358.1.
Whole exome sequencing, bioinformatics, and in silico ANALYSIS
DNA ample was sent to Macrogen (South Korea) (www.macrogen.com) for WES using Novaseq 6000 platform (Illumina San Diego, CA, USA) with 151 bp paired end reads.
In this procedure, DNA was fragmented to create DNA library suite Illumina platform, then fragments were captured to cover all exons, exon/intron junctions, splice sites and flanking intron sequences of all genes (Agilent Sure Select V6 post). All of the fragments were amplified and sequencing was done (mean depth of coverage 100 X for more than 92% 0f the sequences). For bioinformatics analysis, FASTQ file was produced by converting the raw data. In silico, analysis was performed by GATK (Genome Analysis Toolkit) (https://gatk.broadinstitute.org/) and variant calling by BWA (Burrows-Wheeler Aligner) (http://bio-bwa.sourceforge.net/).
In this study, genome alignment and variant detection was based on genome assembly hg19 (hg19, NCBI Build 37), and Picard algorithm was used to mark duplicate reads. Variant filtering was done based on homozygous missense start codon change, splice site, nonsense, stop loss, and indel variants with MAF < 1% in population- based databases such as: db SNP version 147, 1000 genomes project phase 3 database (https://www.internationalgenome.org/), NHLBI GO exome sequencing project (ESP) (https://evs.gs.washington.edu/), exome aggregation consortium (ExAC) (http://exac.broadinstitute.org), and Iranome (http://www.iranome.ir/) and our local exome database (Named GTaC) containing about 1500 exome sequenced samples.
Then, evaluation of the reported variants was done using the following online in silico tools; PROVEAN (http://provean.jcvi.org/), PANTHER (http://www.pantherdb.org/), mutation Taster (http://www.mutationtaster.org/), SIFT (https://sift.bii.a-star.edu.sg/), CADD (https://cadd.gs.washington.edu/) were used to predict their deleterious effect on protein function. In addition, the identified variants were assessed by Human Splicing Finder (HSF) version 3.1 (http://www.ubm.be/hsf/) to predict the effects of them on splice phenomenon. Variant interpretation was accomplished based on the American college of medical genetics (ACMG) (www.acmg.net) guidelines and variant description was based on the Human genome variation society (HGVS) (www.hgvs.org) recommendations.
Variants confirmation
To confirm the WES results and co-segregation analysis, primers were designed by the online web site (http://primer3.ut.ee/), encompassing at least 60 flanking nucleotides of the identified regions of the CDH23 gene (Table 1). Standard polymerase chain reaction (PCR) was applied to amplify the target regions detected by WES. Sanger sequencing was done for the products of PCR.
Results
Clinical findings
There were two affected individuals in the family: a 6-years-old boy (IV3), and a 36-years-old female (III3), that she was his aunt (Fig. 1). The proband was the only offspring of a first cousin consanguineous marriage, which suffered from congenital profound HL. HL in the second patient showed a moderate phenotype, which was different from HL phenotype in the proband. Evaluation of the clinical history showed no other complications in both patients. Retinal examination and the thyroid hormone analysis were performed to relate the HL to Usher or Pendered syndromes, respectively, but the results were normal. Non-syndromic hearing loss was suggested for genetic testing.
WES, Co-segregation analysis
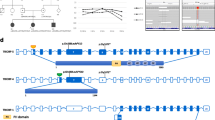
Due to the negative results for GJB2 mutation, WES was applied to define the genetic etiology of HL in the family. Two novel variants were detected in the proband (a homozygous missense variant, c.2961 T > G (NM_022124) in exon 26 of the CDH23 gene, and a heterozygous Splice site variant c.1863-1G > T in OTOGL, (NM_173591)). To confirm the WES results Sanger sequencing was carried out for affected and non-affected individuals of family. The c.2961 T > G variant was detected in the father and mother of the proband as heterozygous state. Proband and his mother were heterozygous for the identified variant in OTOGL too, while the father was homozygous for the wild type variant (Figs. 2 and 3). The second affected member (the proband's aunt) refused to participate in our more examinations.
Bioinformatics analysis and variant interpretations
Deleterious effect of the two identified variants was approved by Bioinformatics tools (Table 2). The effect of variant on splicing was evaluated by HSF (http://umd.be/Redirect.html), and alteration of wild type acceptor site was predicted by the software (Fig. 4). Furthermore, both detected variants have not been reported in the literatures (1000 genomes project phase 3, NHLBI GO ESP, dbSNP version 147, HGMD, ExAC, Clinvar databases and Iranome). Based on the American College of Medical Genetics and Genomics (ACMG) guidelines, c.2961T > G in CDH23 was categorized as likely pathogenic.
Discussions
So far, mutations in more than 150 genes have been reported to cause autosomal recessive non-syndromic sensorineural hearing loss (ARNSNHL). After intellectual disability, non-syndromic hearing loss (NSNHL) is the second cause of inherited disabilities in Iran [36, 37]. Mutations in CDH23 lead to the Usher syndrome type1 (UHS1) or ARNSNHL, type DFNB12 depend on the mutation site and type [38, 39]. Mutations in OTOGL can lead to moderate symmetric, non-progressive form of NSNHL, type DFNB84A [40].
CDH23 protein contains 27 extracellular domains, there are some calcium binding sites (LDRE, DXNDN, and DXD) in these domains, which play essential roles in cell–cell adhesion in the hair cells of the cochlea [41]. Therefore, mutations in the extracellular domains of CDH23 and/or structural changes in these sites can affect calcium binding affinity and protein function. The effect of CDH23: c.2961 T > G variant on amino acid sequence is to change aspartate to glutamate at the position 987 of CDH23 (p. Asp987Glu); this change is occurred in one of the calcium binding sites of the CDH23 protein. It is implied that in the heterozygous state inheritance of the change cannot affect the protein function; so, it cannot lead to hearing loss in the father and the mother of the proband. The homozygous inheritance of the mutation interrupts the function of CDH23 in the proband, and it is the cause of HL due to the loss of function mechanism.
Although, future functional studies are necessary to reveal the real effect of the CDH23:c.2961 T > G variant on protein structure and/or function, mutations in calcium binding sites of the CDH23 lead to ARNSNHL, by disruption of protein function or affecting the calcium-binding affinity in human and mice [42, 43]. Based on the providing reported variant; c.2959G > A (p.Asp987Asn) in the same position and c.2956A > T (p.Lys986Ter) in the same domain of the protein have been led to non-syndromic hearing loss but not the Usher syndrome [38].
The identified variant c.1863-1G > T in the OTOGL gene is a frameshift variant, which interrupts the splicing phenomenon through changing the wild type acceptor site at the junction of intron 18/exon 19. This may lead to a truncated protein, by exon skipping mechanism or longer non-functionally protein, as a result of intron remaining. This variant has been investigated by ET Lim et al., in association with autism spectrum disorders [44].
There are some reports of mutations in OTOGL associated with involvement of visual system [45], but in the current study visual impairment was not observed in the family. In this study, the heterozygous variant in the OTOGL gene was not responsible for the phenotype of the proband. However, it might be the disease etiology in the proband’s aunt but the blood sample was not available for further analysis. Mutations in CDH23 and OTOGL genes are responsible for two different forms of ARNSHL. Also, in some cases of ARNSHL di-genic inheritance of defected alleles (double heterozygosis state) have been led to deafness [35]. There is no evidence for di-genic inheritance of OTOGL/CDH23 genes to cause hearing loss. In this study, the proband’s healthy mother completely rules out the possibility of di-genic inheritance of the cited two variants leading to deafness.
Conclusions
In this study, we reported two novel variants, a homozygous likely pathogenic in the CDH23 gene and a heterozygous splice site in the OTOGL gene.
This study confirmed no ARNSHL phenotype due to double heterozygous for the two mentioned variants. Also, the function of the CDH23 protein is not interrupted in the optical system as a result of homozygous CDH23: c.2961 T > G variant, thereby ruling out the Usher syndrome. Furthermore, double heterozygosis state of the CDH23: c.2961 T > G/OTOGL; c.1863-1G > T variants cannot interrupt the function of optical system. The findings highlighted the importance of genetic counseling and NGS technology in diagnosis of heterogeneous hereditary disorders such as HL.
Data availability
Data availability is the corresponding author's responsibility.
References
Tripathi P, Deshmukh P (2022) Sudden sensorineural hearing loss: a review. Cureus. https://doi.org/10.7759/cureus.29458
Lanzieri TM, Chung W, Flores M, Blum P, Caviness AC, Bialek SR et al (2017) Hearing loss in children with asymptomatic congenital cytomegalovirus infection. Pediatrics. https://doi.org/10.1542/peds.2016-2610
Traylor K, Dodson S, Kralik S, Ho C, Radhakrishnan R (2019) Inner ear anomalies in congenital hearing loss: imaging, genetics, and associated syndromes. Neurographics 9(6):358–372
Wei C-C, Razzak AA, Ghasemi H, Khedri R, Fraase A (2024) Ca2+ binding shifts dimeric dual oxidase’s truncated EF-hand domain to monomer. Biophys Chem 312:107271
van Beeck CE, Engel M, van de Kamp J, Yntema H, Goverts S, Mulder M et al (2019) The etiological evaluation of sensorineural hearing loss in children. Eur J Pediatr 178(8):1195–1205
Zhou Y, Li C, Li M, Zhao Z, Tian S, Xia H et al (2019) Mutation analysis of common deafness genes among 1,201 patients with non-syndromic hearing loss in Shanxi Province. Mol Genet Genomic Med 7(3):e537
Alimardani M, Hosseini SM, Khaniani MS, Haghi MR, Eslahi A, Farjami M et al (2019) Targeted mutation analysis of the SLC26A4, MYO6, PJVK and CDH23 genes in Iranian patients with AR nonsyndromic hearing loss. Fetal Pediatr Pathol 38(2):93–102
de Guimaraes TAC, Arram E, Shakarchi AF, Georgiou M, Michaelides M (2023) Inherited causes of combined vision and hearing loss: clinical features and molecular genetics. Br J Ophthalmol 107(10):1403–1414
Davoudi-Dehaghani E, Bagherian H, DabbaghBagheri S, Mahdieh N, Shirkavand A, Zeinali S (2016) Homozygosity mapping and CDH23 mutation analysis in Iranian deaf families. Hear Balance Commun 14(4):189–193
Dai Z-Y, Sun B-C, Huang S-S, Yuan Y-Y, Zhu Y-H, Su Y et al (2015) Correlation analysis of phenotype and genotype of GJB2 in patients with non-syndromic hearing loss in China. Gene 570(2):272–276
Tabatabaiefar MA, Montazer Zohouri M, Shariati L, Safari Chaleshtori J, Ashrafi K, Gholami A et al (2010) Mutation analysis of GJB2 and GJB6 genes and the genetic linkage analysis of five common DFNB loci in the Iranian families with autosomal recessive non-syndrom. J Sci Islam Republic Iran. 21(2):2
Alipour H, Saudi A, Mirazi H, Kazemi MH, Alavi O, Zeraatpisheh Z et al (2022) The effect of vitamin C-loaded electrospun polycaprolactone/poly (Glycerol Sebacate) fibers for peripheral nerve tissue engineering. J Polym Environ 30(11):4763–4773
Azadegan-Dehkordi F, Bahrami T, Shirzad M, Karbasi G, Yazdanpanahi N, Farrokhi E et al (2019) Mutations in GJB2 as major causes of autosomal recessive non-syndromic hearing loss: first report of c. 299–300delAT mutation in Kurdish population of Iran. J Audiol Otol 23(1):20
Koohiyan M, Hashemzadeh-Chaleshtori M, Salehi M, Abtahi H, Reiisi S, Pourreza MR et al (2018) GJB2 mutations causing autosomal recessive non-syndromic hearing loss (ARNSHL) in two Iranian populations: Report of two novel variants. Int J Pediatr Otorhinolaryngol 107:121–126
Song MH, Lee HK, Choi JY, Kim S, Bok J, Kim UK (2010) Clinical evaluation of DFN3 patients with deletions in the POU3F4 locus and detection of carrier female using MLPA. Clin Genet 78(6):524–532
Riazuddin S, Nazli S, Ahmed ZM, Yang Y, Zulfiqar F, Shaikh RS et al (2008) Mutation spectrum of MYO7A and evaluation of a novel nonsyndromic deafness DFNB2 allele with residual function. Hum Mutat 29(4):502–511
Alasti F, Sanati MH, Behrouzifard AH, Sadeghi A, De Brouwer AP, Kremer H et al (2008) A novel TECTA mutation confirms the recognizable phenotype among autosomal recessive hearing impairment families. Int J Pediatr Otorhinolaryngol 72(2):249–255
Sugiyama K, Moteki H, Kitajiri S-I, Kitano T, Nishio S-Y, Yamaguchi T et al (2019) Mid-frequency hearing loss is characteristic clinical feature of OTOA-associated hearing loss. Genes 10(9):715
Mujtaba G, Bukhari I, Fatima A, Naz SA (2012) C343S missense mutation in PJVK causes progressive hearing loss. Gene 504(1):98–101
Sarmadi A, Nasrniya S, Farsani MS, Narrei S, Nouri Z, Sepehrnejad M et al (2020) A novel pathogenic variant in the LRTOMT gene causes autosomal recessive non-syndromic hearing loss in an Iranian family. BMC Med Genet 21(1):1–9
Alagramam KN, Goodyear RJ, Geng R, Furness DN, Van Aken AF, Marcotti W et al (2011) Mutations in protocadherin 15 and cadherin 23 affect tip links and mechanotransduction in mammalian sensory hair cells. PLoS ONE 6(4):e19183
Usami S-I, Isaka Y, Miyagawa M, Nishio S-Y (2022) Variants in CDH23 cause a broad spectrum of hearing loss: from non-syndromic to syndromic hearing loss as well as from congenital to age-related hearing loss. Human Genet 141(3):903–914
Lin J, Zangi M, Kumar TVNH, Shakar Reddy M, Reddy LVR, Sadhukhan SK et al (2021) Synthetic derivatives of ciclopirox are effective inhibitors of cryptococcus neoformans. ACS Omega 6(12):8477–8487
Gu X, Sun S, Guo L, Lu X, Mei H, Lai C et al (2015) Novel biallelic OTOGL mutations in a Chinese family with moderate non-syndromic sensorineural hearing loss. Int J Pediatr Otorhinolaryngol 79(6):817–820
Pavlenkova Z, Varga L, Borecka S, Karhanek M, Huckova M, Skopkova M et al (2021) Comprehensive molecular-genetic analysis of mid-frequency sensorineural hearing loss. Sci Rep 11(1):22488
Yariz KO, Duman D, Seco CZ, Dallman J, Huang M, Peters TA et al (2012) Mutations in OTOGL, encoding the inner ear protein otogelin-like, cause moderate sensorineural hearing loss. Am J Human Genet 91(5):872–882
Yu S, Choi HJ, Lee JS, Lee HJ, Rim JH, Choi JY et al (2019) A novel early truncation mutation in OTOG causes prelingual mild hearing loss without vestibular dysfunction. Eur J Med Genet 62(1):81–84
Zangi M, Donald KA, Casals AG, Franson AD, Alice JY, Marker EM et al (2022) Synthetic derivatives of the antifungal drug ciclopirox are active against herpes simplex virus 2. Eur J Med Chem 238:114443
Safari H, Zeinali M, Alizadeh P, Mahmoudi D (2024) Topical dexamethason effectiveness combined with surgical intervention in patients suffering from chronic subdural hematoma. Interdiscip Neurosurg 37:101984
Avan P, Le Gal S, Michel V, Dupont T, Hardelin J-P, Petit C et al (2019) Otogelin, otogelin-like, and stereocilin form links connecting outer hair cell stereocilia to each other and the tectorial membrane. Proc Natl Acad Sci 116(51):25948–25957
Iossifov I, O’roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D et al (2014) The contribution of de novo coding mutations to autism spectrum disorder. Nature 515(7526):216–221
Asgharzade S, Tabatabaiefar MA, Mohammadi-Asl J, Chaleshtori MH (2018) A novel missense mutation in GIPC3 causes sensorineural hearing loss in an Iranian family revealed by targeted next-generation sequencing. Int J Pediatr Otorhinolaryngol 108:8–11
Safari H, Jahangiri Babadi A, Alizadeh P, Ajudani R, Hamidi S (2022) Huge unrepaired myelocystocele, progressive sac enlargement in later stages of life: a case report. Br J Neurosurg. https://doi.org/10.1080/02688697.2022.2118235
Harripaul R, Noor A, Ayub M, Vincent JB (2017) The use of next-generation sequencing for research and diagnostics for intellectual disability. Cold Spring Harb Perspect Med 7(3):a026864
Kooshavar D, Tabatabaiefar MA, Farrokhi E, Abolhasani M, Noori-Daloii M-R, Hashemzadeh-Chaleshtori M (2013) Digenic inheritance in autosomal recessive non-syndromic hearing loss cases carrying GJB2 heterozygote mutations: assessment of GJB4, GJA1, and GJC3. Int J Pediatr Otorhinolaryngol 77(2):189–193
Hu H, Kahrizi K, Musante L, Fattahi Z, Herwig R, Hosseini M et al (2019) Genetics of intellectual disability in consanguineous families. Mol Psychiatry 24(7):1027–1039
Koohiyan M, Reiisi S, Azadegan-Dehkordi F, Salehi M, Abtahi H, Hashemzadeh-Chaleshtori M et al (2019) Screening of 10 DFNB loci causing autosomal recessive non-syndromic hearing loss in two Iranian populations negative for GJB2 mutations. Iran J Public Health 48(9):1704
Bademci G, Foster J 2nd, Mahdieh N, Bonyadi M, Duman D, Cengiz FB et al (2016) Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort. Genet Med 18(4):364–371
Ebermann I, Scholl HP, Charbel Issa P, Becirovic E, Lamprecht J, Jurklies B et al (2007) A novel gene for Usher syndrome type 2: mutations in the long isoform of whirlin are associated with retinitis pigmentosa and sensorineural hearing loss. Hum Genet 121(2):203–211
Yariz KO, Duman D, Zazo Seco C, Dallman J, Huang M, Peters TA et al (2012) Mutations in OTOGL, encoding the inner ear protein otogelin-like, cause moderate sensorineural hearing loss. Am J Hum Genet 91(5):872–882
Oroz J, Galera-Prat A, Hervás R, Valbuena A, Fernández-Bravo D, Carrión-Vázquez M (2019) Nanomechanics of tip-link cadherins. Sci Rep 9(1):1–9
de Brouwer AP, Pennings RJ, Roeters M, Van Hauwe P, Astuto LM, Hoefsloot LH et al (2003) Mutations in the calcium-binding motifs of CDH23 and the 35delG mutation in GJB2 cause hearing loss in one family. Hum Genet 112(2):156–163
Wilson SM, Householder DB, Coppola V, Tessarollo L, Fritzsch B, Lee E-C et al (2001) Mutations in Cdh23 cause nonsyndromic hearing loss in waltzer mice. Genomics 74(2):228–233
Lim ET, Raychaudhuri S, Sanders SJ, Stevens C, Sabo A, MacArthur DG et al (2013) Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron 77(2):235–242
Astuti GD, Van den Born LI, Khan MI, Hamel CP, Bocquet B, Manes G et al (2018) Identification of inherited retinal disease-associated genetic variants in 11 candidate genes. Genes 9(1):21
Acknowledgements
Authors appreciate the participation of the patients and their family in the current study. This study was financially supported by the Deputy of Research of the Isfahan University of Medical Sciences (Grant number:197060).
Funding
None.
Author information
Authors and Affiliations
Contributions
Mohammad Amin Tabatabaiefar design the study. Aliasgar Mohammadi, Marziyeh Hoseinzadeh, Sina Narrei and Mohammad Reza Pourreza write the manuscript. Yousof Mohammadi, Mahnaz Norouzi, and Ladan Sadeghian helped in data collection and analysis.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All the procedures performed in the studies involving human participants were in accordance with the ethical standards of the local ethics committee of isfahan university of medical science (IR.MUI.RESEARCH.REC.1397.109), as well as the 1964 Helsinki declaration.
Consent for publication
Not applicable.
Competing interests
Authors declare there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mohammadi, A., Hoseinzadeh, M., Narrei, S. et al. Identification of novel likely pathogenic variant in CDH23 causing non-syndromic hearing loss, and a novel variant in OTOGL in an extended Iranian family. Egypt J Med Hum Genet 25, 102 (2024). https://doi.org/10.1186/s43042-024-00578-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-024-00578-3