Abstract
Background
It remains essential for non-alcoholic fatty liver (NAFLD) patients, to develop a sensitive and specific diagnostic model. Data regarding the use of micro (mi)RNA-34 for NAFLD diagnosis are few. Routine clinical assessment, laboratory tests were done for Egyptian individuals (n = 314) were included (100 healthy individuals and 214 NAFLD patients). Quantification of miRNA-34 was done using real-time PCR. Extremely significant variables were entered into stepwise logistic regression. The diagnostic power of variables was estimated by the area under the ROC (AUC).
Results
MiRNA-34 levels were higher in NAFLD patients than healthy individuals with a significant difference (P< 0.0001). The multivariate analysis was used to evaluate the NAFLD-associated variables (CRP, cholesterol, body mass index (BMI), ALT had p< 0.0001 while mRNA-34 had (p=0.0004). The AUCs (CI) of candidate NAFLD markers were in the order of miRNA-34 0.72 (0.66–0.77) < ALT 0.73 (0.67–0.79) < BMI 0.81 (0.76–0.86) < cholesterol < 0.85 (0.79–0.90) < CRP 0.88 (0.84–0.92). We developed a novel index for discriminating patients with NAFLD named NAFLD Mark. AUC was jumped to 0.98 (0.93–0.99) when five markers were combined. The AUC of NAFLD mark for NAFLD detection was higher than the AUCs of seven common NAFLD indexes (0.44–0.86).
Conclusions
The NAFLD mark is a non-invasive and highly sensitive and specific model for NAFLD diagnosis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Impact statement
Clinical, biochemical, and imaging tools were used for the early diagnosis of NAFLD. A liver biopsy had several disadvantages such as invasive and expensive so there is a need for sensitive and specific tests. The current work aimed to develop a novel model for NAFLD diagnosis using miRNA-34 and routine laboratory parameters
Background
NAFLD is a silent killer disease that is characterized by high hepatic fat aggregation especially aggregation of triglyceride [1]. NAFLD incidence was 25% in the overall population and about 90% among obese patients [2]. The incidence of NAFLD in Egypt is about 16% [3]. NAFLD may lead to chronic liver diseases (hepatic cirrhosis and hepatocellular carcinoma [4]. NAFLD diagnosis is the first step for the evaluation of NAFLD severity [5]. Liver biopsy is the basic tool for NAFLD evaluation, but in routine clinical diagnosis, it has several disadvantages (invasive, risk, and expensive). There is a demand for sensitive, and specific, and non-invasive variables. Clinical and laboratory investigations are used in NAFLD diagnosis [6]. There are several models were developed to evaluate hepatic NAFLD [7]. The fatty liver index (FLI) has an AUC of 0.84 [7]. The hepatic steatosis index (HSI), liver fat score, SteatoTest, and PLALA had AUC of 0.81, 0.86, 80, and 0.86, respectively [8,9,10,11]. The fibrosis-4 (FIB-4) includes age, liver enzymes (AST, ALT), and platelet count [12]. BAAT is based on BMI, age, ALT, and triglycerides [10, 13].
Application of miRNAs (miRNA-29a, miR-34a, and miRNA-122) for NAFLD diagnosis is of particular interest, among which miR-34a, is the most associated with NAFLD development. MiR-34a has a significant role in increasing lipid synthesis and inhibiting mitochondrial fatty acid oxidation in hepatocytes and leading to altered lipid metabolism in NAFLD. miR-34a inhibits gene which regulates catabolism [14, 15]. Therefore, this work aimed to develop a novel model for NAFLD diagnosis using miRNA-34 and clinical and routine laboratory markers. Furthermore, we aim to validate the diagnostic accuracies of seven non-invasive models in comparison with the NAFLD mark for NAFLD diagnosis
Patients and methods
Patients
This case-control study was included healthy individuals (n=100) and NAFLD patients (n=214). All individuals were diagnosed by abdominal ultrasound and FLI score according to Bedogni et al. [16]. Written consent was obtained from all subjects. Exclusion criteria: patients with liver or kidney chronic diseases, and alcohol intake. Full history and clinical examination (BMI and waist circumference) were taken from all patients.
Laboratory investigation
The fasting blood sample (10 ml) was withdrawn from all individuals and divided into three parts: the first part treated with sodium citrate for INR–prothrombin determination, the second part treated with EDTA for complete blood count, and the third part without blood coagulant for evaluation biochemical parameters. Routine biochemical tests were done by automated chemistry analyzer OLYMPUS AU 400 (Olympus America, Pennsylvania, USA). C-reactive protein (CRP) was done using nephelometry kit, CA, USA; alfa fetoprotein (AFP) was measured using CanAg AFP EIA 600-10 and 25 OH D using (EIA-5396; DRG International Inc., Springfield., New Jersey, USA) according to the manufacturer’s instructions.
Determination of the miR-34 level
Serum samples were stored at −80°C until assayed and thawed immediately before the miRNA-34 determination. Firstly, the total RNA was extracted and purified using miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s procedures. cDNA was synthesized by reverse transcription reaction using TaqMan MicroRNA (Applied Biosystems, Foster City, USA) and the thermal cycler (Quanta Biotech). The miR-34 level was amplified from cDNA using TaqMan Universal Master Mix and TaqMan assay (Catalog no: 4427975). The RNU49 was used as a housekeeper gene (Cat no: PN4427975; ID: 001005). All samples were analyzed using the 5 plex Rotor-Gene PCR Analyzer (Qiagen, Germany). The 2ΔΔCt method was conducted for the analysis of gene expression levels using TaqMan microRNA Control Assays RNU49 for normalization purposes. Threshold cycle (CT) was calculated Δ CT value, using the formula [ΔΔ CT = Δ CTtumor−Δ CTnormal]; the fold change was calculated as following [FC= 2- ΔΔ CT] and then log FC was calculated [17].
Statistical analysis
Statistical analysis was achieved using the SPSS statistical and graph pad programs. To analyze the normal distribution of parameters in both groups, the Kolmogorov–Smirnov test was applied. Parametric data were presented as the mean and standard deviation (SD) while non-parametric data were presented as median or interquartile range. Student t test, Mann-Whitney, and chi-square test were used for data analysis. Univariate analysis was done using Student t test, Mann-Whitney, and chi-square test in addition to multivariate analysis were carried out to screen the independent risk factors of NAFLD. The diagnostic value of each variable was assessed by plotting the receiver operating characteristic curve (ROC curve) and determine the area under the curve (AUC). We determined the variable best cut-off value for NAFLD diagnosis (maximum value for the sum of sensitivity and specificity). The diagnostic indexes were expressed as a percentage. Variables with a p value <0.05 at multivariate analyses and high area under ROC were entered into stepwise logistic regression. Common NAFLD indexes (PLALA score [11], FIB-4 [18], BARD [19], NAFLD fibrosis score [12], BAAT [10, 13], AAR [20], and APRI were calculated as original papers [21]).
Results
Patient characteristics
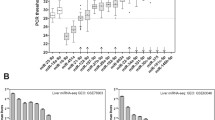
The clinical and laboratory data of the two studied groups were presented in Table 1. MiRNA-34 levels were significantly increased (p< 0.0001) in NAFLD patients than in healthy individuals (Fig. 1A). Using multivariate analysis showed that the increase in miRNA-34 levels was significant (p=0.004). CRP, cholesterol, ALT, and BMI variables were significant (p< 0.0001) associated with the presence of NAFLD.
Diagnostic performance of miRNA-34and candidate’s markers
The diagnostic power of single (CRP, cholesterol, BMI, ALT, and miRNA-34) to diagnose NAFLD was evaluated using area under ROC and presented in Table 2. Variables (CRP, cholesterol, BMI, ALT, and mRNA-34) with a p value <0.05 at multivariate analyses and high AUCs were entered into the stepwise logistic regression. CRP is the most efficient index among clinical and laboratory variables. So, CRP was the best variable to combine with other variables to discriminate against NAFLD patients. Thus, each variable provides independently different information and therefore was expected to increase the diagnostic performances if five variables were combined for the detection of NAFLD. The best linear combinations of blood markers were selected by stepwise multi-discriminant analysis for the development of a novel model (NAFLD mark) based on five markers (CRP, cholesterol, BMI ALT, and miRNA-34). NAFLD mark = [(CRP (mg/L) × 0.025) + cholesterol (mg/dl × 0.005 + BMI (kg/m2) × 0.013 + (ALT (U/L) × 0.008 + miRNA (FC) × 0.002) + 0.044]. The levels of the model in healthy and NAFLD patients were presented in Fig. 1B. The mean [SD] of NAFLD mark studied groups was 1.38 [0.13]; 1.85 [0.25] for healthy individuals, NAFLD with highly significant differences (p < 0.0001). The AUC and diagnostic indexes for the NAFLD mark were presented in Table 2. The NAFLD mark can be used equally effectively in gender without significant difference (p > 0.05).
Evaluation of NAFLD mark vs non-invasive models for NAFLD
The AUC data and the diagnostic performances of seven NAFLD indexes (AAR, PALA, APRI, FIB4, BARD, NAFLD, and BAAT) were compared to NAFLD diagnosis (Table 3, Fig. 2). The NAFLD mark was the most efficient index for NAFLD diagnosis.
Discussion
NAFLD is an over-accumulation of triglyceride in the liver without alcohol consumption. NAFLD can develop chronic liver diseases and hepatic cancer. Liver biopsy is the principal tool with several complications (expensive, invasive with high sampling error). There is a demand for accurate, and non-invasive models and existing non-invasive NAFLD models are inadequate [22]. Diagnosis of NAFLD using various clinical and laboratory data, scoring systems, and imaging methods (abdominal ultrasonography and computed tomography) has been evaluated [23]. The sensitivity of these variables is low. We evaluated several variables to develop the NAFLD mark for NAFLD diagnosis. The NAFLD mark included CRP, cholesterol, BMI, ALT, and miRNA-34. BMI is one of the most variables for obesity evaluation and correlated with the incidence of NAFLD. Higher BMI was associated with the incidence of NAFLD (4–14 fold increase) [24]. BMI was the third diagnostic power variable in the NAFLD mark. This result agrees with the assumption that obesity is the main accountable for NAFLD [2]. The macrophages in fatty cells secrete proinflammatory cytokines such as CRP, and cytokines which impair insulin signaling, inducing insulin resistance. CRP has also been a biomarker of the NAFLD scoring system and a strong predictor of NAFLD. In NAFLD patients, ALT level is accompanying with liver necrosis and liver damage. ALT levels were within normal in 25% of NAFLD patients [25, 26]. ALT was the best predictor of NAFLD had AUC 0.93 with 94% sensitivity and 72% specificity [27]. MiRNA-34 contributes to liver inflammation through an apoptosis pathway and may be used as a biomarker for diagnosing NAFLD [28]. miRNA are small single-stranded RNA (21 to 23 nucleotides) that are responsible for regulating gene expression [29]. In the present study, there was a significant increase (p< 0.0001) in miRNA-34 levels in NAFLD patients than healthy individuals with an AUC of 0.72 for NAFLD diagnosis. The diagnostic power of miR-29a was 0.68 AUC with 61% sensitivity and 82% specificity while miRNA-122 had 0.83 AUC with 75% sensitivity and 82% specificity [30]. Both miRNA-122 and mRNA-34a levels were a significant increase in NAFLD (p<0.001) than normal individuals. miRNA-122 had 92% sensitivity and 85% specificity to discriminate NAFLD from healthy individuals [31]. Simple models have been evaluated including AAR [20], APRI [21], BARD score [19], FIB-4 [18], PALA [11], NAFLD fibrosis score [11], BAAT index [10, 13], FLI, HSI, and NAFLD-liver fat score (NAFLD-LFS). These simple methods do not reflect the mechanism of the NAFLD directly. A developed model for NAFLD diagnosis based on smoking, obesity, hypertension, cholesterol, triglycerides, and ALT had an AUC of 0.81 [32]. For NAFLD diagnosis, FIB-4 had an AUC of 0.81 and APRI had an AUC of 0.73 [33], and the fatty liver index had AUC ranged from 0.81 to 86 while lipid accumulation product had AUC ranged for 0.77–0.92 [34]. Five NAFLD models were evaluated using AUCs. AUCs were in the order of NAFLD liver fat score had AUC of 0.80< hepatic steatosis index had of 0.81< fatty liver index had of 0.83 < triglyceride × glucose model had of 0.90 < visceral adiposity index had 0.92 [35]. Limitations of this study are a single-center study and recommended the validation of NAFLD mark in multi-center studies.
Conclusions
NAFLD mark is a highly sensitive and specific model for NAFLD diagnosis. NAFLD mark showed superior and more accurate diagnostic tool than seven NAFLD indexes for NAFLD diagnosis.
Availability of data and materials
The authors declare that all generated and analyzed data are included in the article.
Abbreviations
- AAR:
-
ALT/AST ratio
- ALT:
-
Alanine aminotransferase
- APRI:
-
Aspartate aminotransferase to platelet ratio index
- AST:
-
Aspartate aminotransferase
- AUC:
-
Area under ROC curve
- BAAT:
-
BMI, age, ALT, triglycerides index
- BARD:
-
Body mass index
- AST:
-
ALT, diabetes index
- BMI:
-
Body mass index
- CI:
-
Confidence interval
- CRP:
-
C-reactive protein
- CT:
-
Threshold cycle
- EDTA:
-
Ethylene diamine tetra-acetic acid
- FC:
-
Fold of change
- FIB-4:
-
Fibrosis index
- FLI:
-
Fatty liver index
- GGT:
-
Gamma-glutamyl transferase
- HSI:
-
Hepatic steatosis index
- INR:
-
International Normalized Ratio
- miRNA:
-
MicroRNA
- NAFLD:
-
Alcoholic fatty liver
- NASH:
-
Nonalcoholic steatohepatitis
- NPV:
-
Negative predictive value
- PLALA:
-
Platelet count, albumin
- PPV:
-
Positive predictive value
- ROC curve:
-
Receiver operating characteristic curve
- SD:
-
Standard deviation
References
El-Zayadi AR (2008) Hepatic steatosis: a benign disease or a silent killer. World J Gastroenterol 14(26):4120–4126. https://doi.org/10.3748/wjg.14.4120
Bellentani S, Scaglioni F, Marino M, Bedogni G (2010) Epidemiology of non-alcoholic fatty liver disease. Dig Dis 28(1):155–161. https://doi.org/10.1159/000282080
Ahmed MH, Noor SK, Bushara SO, Husain NE, Elmadhoun WM, Ginawi IA, Osman MM, Mahmoud AO, Almobarak AO (2017) Non-alcoholic fatty liver disease in Africa and Middle East: an attempt to predict the present and future implications on the healthcare system. Gastroenterology Res 10(5):271–279. https://doi.org/10.14740/gr913w
Younossi ZM (2018) Nonalcoholic fatty liver disease and nonalcoholic steatohepatitis: implications for liver transplantation. Liver Transpl 24(2):166–170. https://doi.org/10.1002/lt.25003
Wong T, Wong RJ, Gish RG (2019) Diagnostic and treatment implications of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Gastroenterol Hepatol (N Y) 15(2):83–89
Abd El-Kader SM, El-Den Ashmawy EM (2015) Non-alcoholic fatty liver disease: the diagnosis and management. World J Hepatol 7(6):846–858. https://doi.org/10.4254/wjh.v7.i6.846
Wong VW, Adams LA, de Lédinghen V, Wong GL (2018) Noninvasive biomarkers in NAFLD and NASH - current progress and future promise. Nat Rev Gastroenterol Hepatol 15(8):461–478. https://doi.org/10.1038/s41575-018-0014-9
Lee JH, Kim D, Kim HJ, Lee CH, Yang JI, Kim W, Kim YJ, Yoon JH, Cho SH, Sung MW, Lee HS (2010) Hepatic steatosis index: a simple screening tool reflecting nonalcoholic fatty liver disease. Dig Liver Dis 42(7):503–508. https://doi.org/10.1016/j.dld.2009.08.002
Kotronen A, Peltonen M, Hakkarainen A, Sevastianova K, Bergholm R, Johansson LM, Lundbom N, Rissanen A, Ridderstråle M, Groop L, Orho–Melander M, Yki–Järvinen H (2009) Prediction of non-alcoholic fatty liver disease and liver fat using metabolic and genetic factors. Gastroenterol 137(3):865–872. https://doi.org/10.1053/j.gastro.2009.06.005
Poynard T, Peta V, Munteanu M, Charlotte F, Ngo Y, Ngo A, Perazzo H, Deckmyn O, Pais R, Mathurin P, Myers R, Loomba R, Ratziu V, FLIP consortium, the FibroFrance-CPAM group, the FibroFrance-Obese group, and the Selonsertib group (2019) The diagnostic performance of a simplified blood test (SteatoTest-2) for the prediction of liver steatosis. Eur J Gastroenterol Hepatol 31(3):393–402. https://doi.org/10.1097/MEG.0000000000001304
Kessoku T, Ogawa Y, Yoneda M, Imajo K, Sumida Y, Eguchi Y, Fujii H, Hyogo H, Ono M, Suzuki Y, Kawaguchi T, Chayama K, Tanaka S, Fujimoto K, Anzai K, Saibara T, Sata M, Itoh Y, Nakajima A, Okanoue T, Japan Study Group of NAFLD (JSG-NAFLD) (2014) Simple scoring system for predicting cirrhosis in nonalcoholic fatty liver disease. World J Gastroenterol 20(29):10108–10114. https://doi.org/10.3748/wjg.v20.i29.10108
Angulo P, Hui JM, Marchesini G, Bugianesi E, George J, Farrell GC, Enders F, Saksena S, Burt AD, Bida JP, Lindor K, Sanderson SO, Lenzi M, Adams LA, Kench J, Therneau TM, Day CP (2007) The NAFLD fibrosis score: a noninvasive system that identifies liver fibrosis in patients with NAFLD. Hepatol 45(4):846–854. https://doi.org/10.1002/hep.21496
Ratziu V, Giral P, Charlotte F, Bruckert E, Thibault V, Theodorou I, Khalil L, Turpin G, Opolon P, Poynard T (2000) Liver fibrosis in overweight patients. Gastroenterol 118(6):1117–1123. https://doi.org/10.1016/S0016-5085(00)70364-7
Ding J, Li M, Wan X, Jin X, Chen S, Yu C, Li Y (2015) Effect of miR-34a in regulating steatosis by targeting PPARα expression in nonalcoholic fatty liver disease. Sci Rep 5(1):13729. https://doi.org/10.1038/srep13729
Liu XL, Pan Q, Zhang RN, Shen F, Yan SY, Sun C, Xu ZJ, Chen YW, Fan JG (2016) Disease-specific miR-34a as diagnostic marker of non-alcoholic steatohepatitis in a Chinese population. World J Gastroenterol 22(44):9844–9852. https://doi.org/10.3748/wjg.v22.i44.9844
Bedogni G, Bellentani S, Miglioli L, Masutti F, Passalacqua M, Castiglione A, Tiribelli C (2006) The Fatty Liver Index: a simple and accurate predictor of hepatic steatosis in the general population. BMC gastroenterol 6(1):33. https://doi.org/10.1186/1471-230X-6-33
Fouda MS, Omran MM, Tarek G, Hady AAWA (2021) Development of a novel panel based on micro-RNAs (21, 29a, 200 and 335) and alpha-fetoprotein as diagnostic biomarkers for hepatocellular carcinoma associated with hepatitis C infection. Arab J Gastroenterol 22(1):28–33. https://doi.org/10.1016/j.ajg.2020.08.004
Sterling RK, Lissen E, Clumeck N, Sola R, Correa MC, Montaner J (2006) M SS, Torriani FJ, Dieterich DT, Thomas DL et al: Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatol 43(6):1317–1325. https://doi.org/10.1002/hep.21178
Harrison SA, Oliver D, Arnold HL, Gogia S, Neuschwander-Tetri BA (2008) Development and validation of a simple NAFLD clinical scoring system for identifying patients without advanced disease. Gut 57(10):1441–1447. https://doi.org/10.1136/gut.2007.146019
Williams AL, Hoofnagle JH (1988) Ratio of serum aspartate to alanine aminotransferase in chronic hepatitis. Relationship to cirrhosis. Gastroenterol 95(3):734–739. https://doi.org/10.1016/S0016-5085(88)80022-2
Wai CT, Greenson JK, Fontana RJ, Kalbfleisch JD, Marrero JA, Conjeevaram HS, Lok AS (2003) A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatol 38(2):518–526. https://doi.org/10.1053/jhep.2003.50346
Piazzolla VA, Mangia A (2020) Noninvasive Diagnosis of NAFLD and NASH. Cells. 9(4):1005. https://doi.org/10.3390/cells9041005
Sumida Y, Nakajima A, Itoh Y (2014) Limitations of liver biopsy and non-invasive diagnostic tests for the diagnosis of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J Gastroenterol 20(2):475–485. https://doi.org/10.3748/wjg.v20.i2.475
Fan R, Wang J, Du J (2018) Association between body mass index and fatty liver risk: a dose-response analysis. Sci Rep 8(1):15273. https://doi.org/10.1038/s41598-018-33419-6
Ma X, Liu S, Zhang J, Dong M, Wang Y, Wang M, Xin Y: Proportion of NAFLD patients with normal ALT value in overall NAFLD patients: a systematic review and meta-analysis. BMC Gastroenterol 2020 14;20(1):10 .
Preuss HG, Kaats GR, Mrvichin N, Bagchi D, Preuss JM (2019) Circulating ALT levels in healthy volunteers over life-span: assessing aging paradox and nutritional implications. J Am Coll Nutr 38(8):661–669. https://doi.org/10.1080/07315724.2019.1580169
Martin-Rodriguez JL, Gonzalez-Cantero J, Gonzalez-Cantero A, Arrebola JP, Gonzalez-Calvin JL (2017) Diagnostic accuracy of serum alanine aminotransferase as biomarker for nonalcoholic fatty liver disease and insulin resistance in healthy subjects, using 3T MR spectroscopy. Medicine 96(17):e6770. https://doi.org/10.1097/MD.0000000000006770
Muangpaisarn P, Jampoka K, Payungporn S, Wisedopas N, Bunchorntavakul C, Tangkijvanich P, Treeprasertsuk S (2017) Serum microRNA-34a is potential biomarker for inflammation in nonalcoholic fatty liver disease. Asian Biomedicine 10(2):163–171. https://doi.org/10.5372/1905-7415.1002.478
Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA, Sanyal AJ (2008) Nonalcoholic steatohepatitis is associated with altered hepatic microRNA expression. Hepatol 48(6):1810–1820. https://doi.org/10.1002/hep.22569
Jampoka K, Muangpaisarn P, Khongnomnan K, Treeprasertsuk S, Tangkijvanich P, Payungporn S (2018) Serum miR-29a and miR-122 as potential biomarkers for non-alcoholic fatty liver disease (NAFLD). MicroRNA 7(3):215–222. https://doi.org/10.2174/2211536607666180531093302
Hendy OM, Rabie H, El Fouly A, Abdel-Samiee M (2019) The circulating micro-RNAs (-122, -34a and -99a) as predictive biomarkers for non-alcoholic fatty liver diseases. Diabetes Metab Syndr Obes 12:2715–2723. https://doi.org/10.2147/DMSO.S231321
Abd El-Wahab EW, Zein El-Abedin RA (2020 Dec) Ahmed WM. Shatat HZ: Validation of a non-laboratory based screening tool for predicting non-alcoholic fatty liver disease in an Egyptian setting. am J Med Sci 360(6):662–677. https://doi.org/10.1016/j.amjms.2020.06.020
Baikpour M, Ozturk A, Dhyani M, Mercaldo ND, Pierce TT, Grajo JR, Samir AE: Portal venous pulsatility index: a novel biomarker for diagnosis of high-risk nonalcoholic fatty liver disease. AJR Am J Roentgenol., 214(4):786-791.
Kozłowska-Petriczko K, Wunsch E (2021) Diagnostic accuracy of non-imaging and ultrasound-based assessment of hepatic steatosis using controlled attenuation parameter (CAP) as reference. J Clin Med 10(7):1507. https://doi.org/10.3390/jcm10071507
Fedchuk L, Nascimbeni F, Pais R, Charlotte F, Housset C, Ratziu V (2014) Performance and limitations of steatosis biomarkers in patients with nonalcoholic fatty liver disease. Aliment Pharmacol Ther 40(10):1209–1222. https://doi.org/10.1111/apt.12963
Acknowledgements
Not applicable
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
OM and MA conceived and designed experiments. The authors contributed to the analysis and/or interpretation of the data equally. OM and MA drafted the manuscript. OM and MA revised it critically for important intellectual content. The authors have read the manuscript and approved the final submission.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Faculty of Medicine, Minia University, Minia, Egypt (Code # IRB 19:1/2021). NAFLD mark: An accurate model based on microRNA-34 for diagnosis of non-alcoholic fatty liver disease patients. Informed written consent was signed by all patients in compliance with the ethical guidelines of the 1975 Declaration of Helsinki.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mohamed, A.A., El-Demery, A., Al-Hussain, E. et al. NAFLD mark: an accurate model based on microRNA-34 for diagnosis of non-alcoholic fatty liver disease patients. J Genet Eng Biotechnol 19, 157 (2021). https://doi.org/10.1186/s43141-021-00257-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43141-021-00257-5