Abstract
In Poland, red deer, roe deer and elk cause great damage to pine stands, and pine is the predominant forest-forming tree species in Polish forests. Pine seedlings planted adjacent to agricultural fields face heightened risk of damage from nocturnal foraging by game animals, which retreat to forested areas during the day. This damage creates openings for fungal entry, particularly by concerning plant pathogens, such as Fusarium species, which are prevalent in agricultural settings. This study aimed to analyze the mycobiome of pine seedlings damaged by forest animals, focusing on the presence of concerning pathogens in infested pine seedlings collected from ten forest plantations at various distances from rural areas. Samples of pine seedlings bitten by forest animals were taken from 10 forest plantations at different distances from rural areas. Selected diversity indices were determined to compare the fungal populations. Mycological analysis of the gnawed pine shoots using culture and qPCR methods revealed 746 fungal colonies. The most frequently isolated species were: Fusarium avenaceum, Alternaria alternata, Botrytis sp., Cladosporium cladosporioides, representatives of the genus Penicillium, Rhizopus nigricans and Fusarium oxysporum, which were found in samples from all study plots and are thus ubiquitous. Species that are typical pathogens of agricultural crops, such as Fusarium avenaceum, F. culmorum, F. poae, F. sporotrichioides and F. tricinctum, were also isolated from damaged seedling tissue in forest plantations neighbouring agricultural fields. Spearman’s rank correlation analysis showed that the importance of secondary pathogens, represented by species of the genus Fusarium, decreased with increasing distance from agricultural fields, while the importance of primary pathogens increased. Younger pine seedlings exhibited lower species diversity and a higher prevalence of specific secondary pathogen species, coupled with heightened susceptibility to infection compared to older seedlings when subjected to tissue damage by forest animals. These findings underscore not only variations among plantations in proximity to agricultural fields but also suggest potential inter-land spread of fungi, emphasizing the need for integrated management strategies across agricultural and forestry domains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ecological function of game animals in forest plantations
Scots pine (Pinus sylvestris L.) is the most widespread conifer in the pine family (Pinaceae) and among the gymnosperms that grow in the northern hemisphere. It is a valuable and commercially important timber tree in Europe. According to Palmer and Truscott (2003), browsing of pine seedlings by game is an important factor limiting the natural regeneration of pine forests. Conservation considerations, such as the ban on elk hunting in Poland, are leading to increasing game pressure in agriculture and forestry. In Poland, there are an estimated 282,000 red deer (Cervus elaphus L.), 920,000 roe deer (Capreolus capreolus L.) and 31,000 elk (Alces alces L.) on an area of around 9 million hectares of forest (Poland, 2021). While there is no data on fallow deer (Dama dama L.), they are known to occur in both agricultural and forested areas. Deer damage in agricultural crops and young plantations is an important economic problem and silvicultural challenge for foresters, as these animals damage plant organs such as bark, buds, shoots and leaves and also pull seedlings out of the ground (in the case of material from containerised nurseries) (Pfeffer et al., 2021; Ramos et al., 2006).
The occurrence of pathogenic fungi in pine cultivations
Pine trees are usually grown from seed, and the most important time for disease control is during the early growth stages in plantations, where many forest animals find food. Fungi of the genera Fusarium, Alternaria, Botrytis, Rhizoctonia, Cylindrocarpon and the fungus-like Phytophthora or Pythium can infect emerging seedlings and the roots of young plants in the first years of growth and are thus potential pathogens that can be transferred from agricultural land to forest ecosystems. Fungal infestations are often lethal and can severely limit plant growth or even prevent the establishment of seedlings (Harrington, 1977).
Risk of transmission of pathogens between ecosystems
Although a direct link between animal transmission of fungi to seedlings has not yet been proven, it cannot be excluded. Insects are often mentioned as vectors of plant pathogenic fungi in the literature (Lee, 2023; Scarlett et al., 2014; Yamoah et al., 2011), but there is no information on the role of animals as plant pathogen vectors, which may be indirectly suggested in this study (but was not the aim of the study). It is known, for example, that birds can transmit numerous pathogens on feathers over short and long distances Malewski et al. (2019). If they eat agricultural seeds or fruits they can also transfer pathogens between both ecosystems (Mancini & Romanazzi, 2014; Zhang et al., 2019).
We suspect that the first invaders of forest plantations came from agricultural crops and that, in addition to dispersal by wind or water, forest animals are also involved, probably transmitting the spores via the hooves (with the soil) as well as via the fur. Many pathogens previously found on agricultural plants (Fusarium avenaceum, F. graminearum, F. poae or F. culmorum) have the potential to infect forest seedlings.
Indeed, Roques et al. (2015) found that, after planting European indicator trees in East Asia as a new method to identify potential pest invaders, most of the identified pathogen species originated from the surrounding agricultural crops and fruit trees and not from the forests. In addition to F. oxysporum, which is a known pathogen of forest seedlings, many other agricultural pathogen species could pose additional threats as they can also infect forest tree seedlings (Olaizola et al., 2023).
We do not know whether Fusarium species typical of agricultural (non-forest) crops were introduced from neighbouring rural crops in the past, but it seems likely. However, where they have appeared in forest plantations, their further development must be monitored for possibility of disease symptoms.
In 2022, in the Polish Warminsko-Mazurskie Voivodeship (the area where this study was conducted), cereals were grown on an area of approximately 420 thousand hectares, with the most important species such as wheat and triticale grown on 184 thousand hectares and 71 thousand hectares, respectively. Another important crop in the Warminsko-Mazurskie Voivodeship is winter oilseed rape, which is grown on 99 thousand hectares, and maize, which is grown on almost 60 thousand hectares (GUS, 2023). In the immediate vicinity of forest crops, winter wheat and winter oilseed rape were grown on agricultural land, while in more distant forest crops, wheat and oilseed rape as well as maize and spring barley were grown in addition to the above-mentioned crops.
Plant pathogens, including Fusarium and saprotrophic fungi
The species F. avenaceum, F. graminearum, F. culmorum and F. tricinctum are among the most important pathogens of Fusarium Head Blight (FHB) in cereals (Okorski et al., 2022). In addition, as polyphagous species, they can also cause diseases of many crops, leading to an accumulation of mycotoxins that are dangerous to human and animal health (Okorski et al., 2017). The disease occurs practically every year with varying intensity, depending on the current weather conditions and the availability of inoculum in the environment (Okorski et al., 2022). However, it should be noted that different Fusarium species have been identified in the same field, in the same ear and even in asymptomatic grain (Kulik, 2008).
For this reason, in this article we focus on the genus Fusarium, which is a major threat to agricultural crops and is also known as a pathogen in forestry (especially in the juvenile stage). In addition, other studies (Neupane et al., 2013) have shown that the rhizosphere of plants can be colonised by other species in the Fusarium genus, including: F. avenaceum (Fr.) Sacc., F. culmorum (Wm.G. Sm.) Sacc., F. oxysporum Schltdl., F. tricinctum (Corda) Sacc. and Fusarium graminearum Schwabe. These species cause red ear rot in maize, which leads to a high accumulation of mycotoxins in this plant material such as deoxynivalenol (DON) and zearalenone (ZEA) (Bocianowski et al., 2020; Czarnecka et al., 2022).
We hypothesise that Fusarium fungi (F. avenaceum, F. graminearum, F. culmorum or F. tricinctum) commonly found in agricultural crops represent a high infection potential, threatening pine plantations under stress conditions as a consequence of bites by forest animals such as deer or roe deer. In agriculture, for example, these species are among the most important pathogens causing Fusarium Head Blight (FHB) in all cereals (Okorski et al., 2022), but in addition, they can also cause diseases of other plants, including forest trees, e.g. oak bud blight (Wit et al., 2015) or damping off of pine seedlings or lethal diseases in mature trees (Drenkhan et al., 2020; Elvira-Recuenco et al., 2019). Only a few studies (Arhipova et al., 2015; Šnepsts et al., 2022) describe an interaction between wild browsing and the occurrence of diseases in forest crops. Debarking led to a significantly greater reduction in resistance to intrinsic wood damage (primary decay) compared to root rot, while it had a similar effect on resistance to lethal (secondary) damage (Šnepsts et al., 2022). The study by Arhipova et al. (2015) showed that the species Sarea difformis (Fr.) Fr and Peniophora pini ((Schleich.) Boidin), both of them were most frequently isolated from wounds. On the other hand, there is a lack of work describing the effects of shoot gnawing on the general physiological condition and health of pine seedlings.
The aim of the study was to analyse the structure and species composition of the mycobiome of pine seedlings damaged by forest animals in comparison to undamaged seedlings.
Materials and methods
Field study
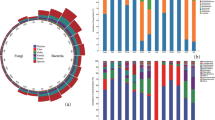
The material for the study (the top shoots of pine seedlings bitten by forest animals) (Fig. 1) was collected on 27 September 2022 in the Uklanka forestry area, within the Strzałowo forest district (northeastern Poland). Ten sample plots (Fig. 2, Table 1) were selected based on the predominance of Scots pine in the species composition.
Isolation of fungal communities from pine shoots with bite symptoms
Isolation of the fungal communities, both of pathogens and endophytes of the gnawed tree tissue, was carried out in the laboratory of the Department of Entomology, Phytopathology and Molecular Diagnostics of UWM Olsztyn. Pine shoots with feeding symptoms (necrotic tissues) were collected in the defined test plots, from 20 shoots per plot, from 2- to 7-year old seedlings (Table 1). The control group consisted of seedlings without bite symptoms. Five seedlings from each test plot were combined into one sample. The seedlings with visible bites were cut with a scalpel into 2–3 mm fragments taken from damaged tissue (per plot, 7 fragments were placed on 15 Petri dishes, giving in total a hundred and five fragments per plot).
In the laboratory, the collected shoots were mechanically cleaned of organic contaminants and the pine needles were removed, leaving the top of the main shoot. The boundary between necrotic and healthy tissue was left for further examination, which was cut with a scalpel into 5 mm fragments (having removed the bark). The plant material was then divided into two parts to obtain material for molecular studies. Half of the collected plant material was surface sterilised in 70% ethanol for 30 seconds, followed by 0.1% sodium hypochlorite solution (NaClO), to isolate the fungal communities, and the other half was used to perform molecular analyses. Finally, the samples were rinsed three times for 30 seconds each in sterile water.
Five fragments with bite symptoms were lined up on a plate, and the samples for each test variant were lined up on 20 Petri dishes with Potato Dextrose Agar (PDA) medium. The plates were stored in a growth chamber at 23 °C with a cycle of 12 hours of darkness and 12 hours of UV light to promote fungal growth and sporulation. For 10 days, the growing fungal colonies were checked daily and each new hyphae was transferred to new sterile Petri dishes on PDA medium.
Fourteen days after removal from the culture system, the colonies were transplanted onto new dishes with PDA medium. The fungal colonies were sequentially assigned to a species/genus based on morphological characteristics such as mycelium type, pigmentation, shape of mycelia, size of mycelia, type of mycelium formation and presence of spore forms. Where possible, fungal colonies were identified to genus and species level based on morphological characteristics such as spore shape and size and type of sporulation. Microscopic examination was performed using a light Olympus CX40 (Tokyo, Japan) microscope to identify the fungi using the available key (Ellis, 1971; Gerlach et al., 1982; Leslie & Summerell, 2006; Watanabe, 2010).
DNA extraction from pine shoots
Total genomic DNA was extracted from the frozen tissue of a single individual with bite symptoms (five replicates per research variant) using the Maxwell 16LEV Plant DNA Kit (Promega, Madison, WI, USA). The quality of the DNA was checked on a 1% (w/v) agarose gel stained with 0.5 μg/mL ethidium bromide. The concentration and purity of the DNA samples were determined spectrophotometrically (NanoDrop ND 2000C UV-Vis spectrophotometer; Thermo Fisher Scientific, Waltham, MA USA) and with the Florymeter (Qubit 2, Thermo Fisher Scientific, Waltham, MA USA). Only DNA matrices with good quality parameters were selected for further analyses. For the qPCR analyses, the pine samples were normalised by bringing them to an equal concentration of 1 ng total DNA based on fluorimeter measurements.
The reference material for the qPCR analyses was gDNA from isolates: F. avenaceum (isolate p12.56.2016), F. graminearum (123/23/2017) and Phytophthora cactorum (Lebert & Cohn) J. Schröt. (isolate 158), isolated from mycelium scraped with a sterile scalpel from the surface of 14-day-old PDA cultures. Cultures were ground (homogenised) with a pestle and mortar in liquid nitrogen according to the protocol using a Maxwell® 16 FFS Nucleic Acid Extraction System. Serial dilutions of gDNA from each isolate were prepared for qPCR at concentrations ranging from 1 ng to 1 fg gDNA in 1 μL. All isolates used in this study were obtained from the collection of the Department of Entomology, Phytopathology and Molecular Diagnostics of the Faculty of Agriculture and Forestry of the University of Warmia and Mazury in Olsztyn, Poland.
Detection of Fusarium and Phytophthora using qPCR technology
The qPCR method (for quantification of pathogen DNA) was used to test whether fungal genotypes typical of agricultural crops were present in large numbers in pine seedlings exposed to biting stress. The material for qPCR analysis consisted of matrix DNA from extracted from the sampled pine seedlings. The qPCR of 30 samples (10 samples × 3 independent isolates per matrix) and control samples (DNA isolated from frozen shoots at −80 °C without biting symptoms) were analysed in three wells (qPCR results showed the mean with replicate), and all matrices were analysed for all fungal species. For each gDNA template analysed, the qPCR reaction was performed in triplicate. The study was performed with Taq Man primers and probes that allowed detection of the following genotypes:
-
F. avenaceum/F. tricinctum (Esyn1): Avetric F: GGCYTTCCTGCGAACTTG; Avetric R: AGC AGT CGA GTT CGTCAACAGA; Avgetric Probe: Fam-CCGTCG AGT CCT CT -Mgb (Kulik et al., 2011)
-
F. culmorum/F. graminearum (Tri5): Tri5 forward: TCT TAA CACTAG CGTGCGCCT TC; Tri5 reverse: CAT GCCAAC GAT TGT TGT TTG GAG GGA; FGTri5-probe: Fam-AACAAGGCTGCCCACCACTTTGCT CAG CCT-Tamra (Vegi & Wolf-Hall, 2013).
-
Phythophthora spp. FITS _15Ph: TGCGGAAGGATC ATT ACC ACC; RITS_279Ph: GCGAGCCTAGACATCCAC TG; All_PhTaqMan probe: Fam-TTGCTATCT ATT AAA AGCA-Mgb (Sikora et al., 2012).
The study was performed on a 7500 FAST real-time PCR system (Applied Biosystem, Thermo Fisher Scientific, Waltham, MA USA) using TermoFisher Scientific reagents containing a qPCR reaction mixture of 20 μL, including: 10.0 μL TaqMan Universal PCR Master Mix (Applied Biosystems, USA), 10 pM of each primer, 10 pM of each primer labelled with FAM at the 5′ and 6′ ends and TAMRA quencher at the 3′ end, 4.0 μL deionised water and 5 μL DNA. The amplification conditions were as follows: initial denaturation at 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 15 s, primer annealing at 60 °C for 15 s and strand synthesis at 72 °C for 1 min.
The amount of DNA isolated from each genotype of Fusarium or Phytophthora was determined using calibration curves (Table 6) and the calibration curves of Livak and Schmittgen (2001) and Pfaffl (2001) with some modifications.
Statistical analysis of the collected data
In order to compare the DNA levels of the analysed plant pathogens in plant tissues, analysis of variance (ANOVA) was performed together with the one-way Tukey test (HSD), and values of p < 0.01 were considered significant.
To determine the relationship between the results of the cultural method for fungal identification and the qPCR tests, a linear regression analysis was performed, and the Pearson correlation coefficient (R) was determined. To determine the relationship between biodiversity indices of trophic fungal endophytic groups and the average distance from fields and age of pine seedlings, the Spearman rank correlation method was used, with a significance level of p ≤ 0.05. STATISTICA 13.3 software (TIBCO Software Inc., n.d.) was used for the mathematical analyses.
Data analysis of the trophic group of fungi
Statistical analyses were carried out to determine, among other things, the effect of the distance from agricultural fields on the occurrence of endophytes - secondary pathogens such as Fusarium - which are typical of agricultural ecosystems. Biodiversity indices were used to quantify the relationships between abundance, distribution preference and composition of the studied fungal communities (Table 2).
Once classified, the species identified in the mycobiome of the analysed seedlings were assigned to the following trophic groups:
-
endophyte primary pathogens (EPP) - these are the organisms that cause the disease (Reid, 2006)
-
endophyte secondary pathogens (ESP) - these are other organisms that can either cause disease or simply infest and feed on dead plant tissue caused by pests, disease or other injury. Secondary pathogens can occur as parasites that kill living tissue or as saprotrophs (Reid, 2006)
-
endophyte saprotrophs (ES) - organisms that utilise non-living/dead organic material as food (Reid, 2006).
Spearman rank regression analysis was used to analyse the effect of distance from agricultural fields and the age of seedlings on the biodiversity indices for each individual trophic group.
Results
Identification of isolates
The presence of primary pathogens in pine on the study plots was as follows:
-
Pine shoot dieback (Sydowia polyspora (Bref. & Tavel) E. Müll.) was observed on all plots except 6.
-
Pine twisting rust (Melampsora populnea (Pers.) P. Karst.) occurred in older plots, in plots 2,3,5,7 and 10.
-
Shoot dieback (Sphaeropsis sapinea (Fr.) Dyko & B. Sutton) occurred in all plots except 1, 6, 8 and 9.
Mycological analysis of the gnawed pine shoots using the culture method revealed 759 fungal colonies (Table 3). The most frequently isolated species were Fusarium avenaceum, Alternaria alternata (Fr.) Keissl, Botrytis cinerea Pers., Cladosporium cladosporioides (Fresen.) de Vries, representatives of the genus Penicillium, Rhizopus nigricans Ehrenb., and Fusarium oxysporum, which were found in samples with biting symptoms from all study plots. Fungal isolates found on the shoots of the control seedlings included A. alternata, Aureobasidium bolleyi (R. Sprague) Arx., Cladosporium cladosporioides, Cylindrocarpon obtusisporum (Cooke & Harkn.), and Mucor circinelloides Tiegh.
The biodiversity indices of the tropic groups of the fungal endophytes – dividing endophytes into primary pathogens (EPP), secondary pathogens (ESP) and saprotrophs (ES) – showed that saprotrophic species were predominant at all tested plots [Rf ranged from 1.71–7.25] (Table 4). Seedlings without biting symptoms (control group) only had saprotroph endophytes [Rf = 1.71], with no primary or secondary pathogens present. A high relative frequency index for primary pathogens was found for plots 1 (2.11), 4 (1.84), and 10 (1.84). The relative frequency index of the secondary pathogen group was higher, ranging from 1.58 on plot 4 to 3.69 on plot 10. The dominance index [Y] was highest for the saprotrophic populations at practically all the sites analysed, and also for the control group. Only at site 10 were similar values determined for the index [Y] for the saprotrophs endophytes [Y = 0.17] and secondary pathogen endophytes group [Y = 0.14] (Table 4).
The species richness of the primary and secondary endophyte pathogens was very similar and ranged between 2 and 6 species for EPP and 2–6 species for ESP depending on the site, while the saprotrophic communities were more diverse with 5–15 species.
The higher diversity of the saprotrophic fungal endophytes was confirmed with the Shannon-Weiner [H′] and Margalef [D′] indices, whose values were higher for the saprotrophic populations at virtually all sites (Table 4). The dominance index [λ] confirmed the dominance of the endophyte saprotrophs for sites 1–5, and 7, and the seedlings of the control group. For sites 6 and 8–10, the higher values of the [λ] index were found for the endophyte secondary pathogens group. The group of secondary pathogens consisted mainly of Fusarium species that are typical agricultural plant pathogens, such as F. avenaceum, F. culmorum, F. poae, F. sporotrichioides and F. tricinctum.
Molecular analysis
The qPCR results show that the most important fungal species in agricultural crops, i.e. Fusarium spp. – the most important pathogen causing Fusarium Head Blight (FHB) of all cereals in Poland – were detected in infested pine seedlings (Okorski et al., 2019). The genotype of F. avenaceum/F. tricinctum was found in samples from all study plots except the control (Fig. 3).
Results of the quantitative evaluation of the prevalence of the genotype of F. avenaceum/F. tricinctum in bite material from forest plantations according to location [pg DNA]. Different capital letters show significant differences by Tuckey’s test at p < 0.01; 1–10- mark the sampling areas *C- control group
The highest DNA concentrations were found in samples from plot 9 (146.8 pg) and plot 6 (92.5 pg). The lowest DNA concentration of the analysed genotype was found in samples from plot 10 and plot 3 (Fig. 3).
The F. culmorum/F. graminearum genotype was most abundant in plots 1, 10 and 7, where 29.3, 18.3 and 13.8 pg DNA were detected, respectively. Trichothecene-producing Fusarium fungi were not found on the seedlings in plots 2, 5, 6, 8 and 9 or on the control seedlings without feeding symptoms (Fig. 4).
The presence of oomycete pathogens was only detected in samples from sites 2 and 9, which contained 20.6 pg and 1.5 pg of DNA, respectively (Fig. 5).
Mathematical analysis was then used to determine whether the results of the mycological analysis and the results obtained by qPCR were compatible. For this purpose, a linear regression analysis was performed, which showed a positive correlation between the number of cultures of F. avenaceum and F. tricinctum and the amount of DNA of the two species detected by quantification of the ESYN1 gene (Fig. 6).
Linear regression between the amount of gDNA of the genotype of F. avenaceum and F. tricinctum and the sum of F. avenaceum and F. tricinctum isolates from bite material in forest cultivation. The regression formula and the correlation coefficient R are shown in the figure. For the regression slope coefficient p < 0.01
The result of the analysis showed a positive correlation, which is confirmed by the value of Pearson’s linear correlation coefficient (R = 0.90). Similarly, a linear regression analysis was performed in relation to the number of isolates of F. culmorum and F. graminearum identified, and the amount of DNA of both species expressed by the amount of TRI5 gene (Fig. 7). The analysis showed a positive correlation between the variables, as evidenced by the high Pearson correlation coefficient of R = 0.95.
Linear regression between the amount of F. culmorum/F. graminearum genotype gDNA and the sum of F. culmorum and F. graminearum isolates from bite material in forest cultivation. The regression formula and the correlation coefficient R are shown in the figure. For the regression slope coefficient p < 0.001
Influence of distance from fields and age of seedlings on fungal communities
Statistical analysis showed that the distance of agricultural fields from the investigated forest crops has a significant positive influence on the prevalence of the most important fungal pathogens on bitten pine seedlings. The analysis showed that as the distance from the agricultural fields increased, the importance of the primary pathogens on the bitten shoots of the pine seedlings increased. Plot No. 1, which is located 800 m away from fields that are used for growing crops, was determined to have the highest degree of species richness [S] index and 19% percentage of endophytes were from primary pathogen group (Table 3). Four fungal species were identified from the primary pathogen group: Lophodermium pini-mugonis, Diplodia pinea, Ilyonectria destructans, and Botrytis cinerea. The percentage of primary pathogens to all isolated fungi dropped as this distance reduced. Plots 3 and 7 (which immediately border agricultural fields) and 9 (which is 200 m away from an agricultural field) revealed 11%, 7%, and 13% of the total number of primary pathogens, respectively. One or two species from this group were discovered in these plots (Table 4).
The above observations were confirmed by calculating the values of the coefficients Spermans rank correlation for relative frequency [Rf] and dominance [Y] of primary pathogens (R = 0.63 and R = 0.56, respectively) (Table 5).
An inverse relationship (negative correlation) was found for the secondary pathogens. The analysis showed that the lower the distance to the agricultural fields, the higher the values of the biodiversity indices describing the fungal communities categorised as secondary pathogens. The Spearman’s Rank correlation coefficients determined were for Relative frequency: R = -0.57, Dominance: R = − 0.58, Species richness: R = -0.68, Maraglef index: R = -0.59, Shannon-Weiner index: R = -0.77 (Table 5). The group of secondary pathogens included Fusarium species, whose infection level was determined by qPCR. Therefore, a linear regression analysis was performed to determine the strength of the relationship between the infection level of gnawed shoots (by Fusarium spp.) and the distance between the studied pine stands and the agricultural fields (Fig. 8(a)). The linear regression analysis also showed a weak negative correlation between the distance to the agricultural fields and the DNA levels of the tested Fusarium genotypes (qPCR method) (R = -0.2).
Linear regression between the amount of Fusarium species DNA and the average distance from agricultural fields (a) and age of pine seedlings (b). The formula of the regression line and the correlation coefficient R are shown in the subfigures. The p value of the regression coefficient is not statistically significant for the data shown in subfigure (a), for subfigure (b) p = 0.02
The mathematical analysis showed that older pine seedlings with browsed shoots had higher species diversity of secondary pathogens. This was indicated by a Maraglef index R-value of 0.71 and a Shannon-Weiner index R-value of 0.72 (Table 5). On the other hand, younger seedlings were characterised by a lower species diversity and a dominance of certain secondary pathogen species. This was also confirmed by a linear regression analysis of the amount of Fusarium fungal DNA determined by qPCR and the age of the deer-infested seedlings.
In addition, analysing the effects of the age of pine seedlings in each study plot showed that younger pine seedlings were more infected with Fusarium compared to older ones. This relationship was confirmed by a linear regression analysis (Fig. 8b), which showed a Pearson’s linear correlation coefficient of R = −0.48 (p = 0.02).
Discussion
Mycobiome diversity within plots
Virtually all of the study plots contained the following species: A. alternata, B. cinerea, F. avenaceum, F. oxysporum, Penicillium sp. and Rhizopus nigricans (Table 3). These species represented all trophic groups of endophytes, including primary pathogens, secondary pathogens, and saprophytes. In addition, the species C. cladosporioides, I. destructans and representatives of the genus Aureobasidium were relatively abundant on the gnawed seedlings of most of the analysed experimental plots. The other species on the seedlings in the analysed plots were only sparsely represented.
The seedlings undamaged by forest animals, which formed the control group, showed an undifferentiated mycoflora with 13 fungal isolates representing five taxa. The dominant species was A. alternata, and there were also some representatives of endophytes: A. bolleyi, C. cladosporioides, Cylindrocarpon obtusisporum and Mucor circinelloides (Table 3).
Finally, it should be noted that other dangerous root pathogens such as Heterobasidion annosum in area 205j and Armillaria ostoye (its rhizomorphs were found on a dead tree standing at the edge of the ecotone in section 20c) were also present. However, Nectria cinnabarina was found in all plots, mainly on dead wood (branches, shoots) lying on the litter, but in some cases it also colonised dead branches of standing trees, such as willows Salix caprea L. All the fungi mentioned above can also be exchanged between plots, which will be a separate research topic in the future.
The species F. avenaceum, F. graminearum, F. culmorum and F. tricinctum, which were detected on pine seedlings in our studies, are among the most important agricultural pathogens, causing Fusarium Head Blight (FHB) in cereals (Okorski et al., 2022). However, as polyphagous species, they can also cause diseases in many other crop plants, leading to an accumulation of mycotoxins that are dangerous to human and animal health (Okorski et al., 2017).
In relation to pine, one study by Davydenko et al. (2018) found that indigenous strains of different Fusarium species isolated from wheat plants and wheat grains (F. culmorum, F. graminearum, F. oxysporum, F. poae, F. tricinctum) showed weak pathogenic properties against pine seedlings from different Polish areas of origin. However, individual strains of the same Fusarium species may differ in their infectivity, possibly due to their ability to produce mycotoxins. Within the same Fusarium species, there are different strains with the ability to synthesize different mycotoxins (Buśko et al., 2014). The possibility of infection of pine seedlings is indicated by an earlier study by Okorski et al. (2019), which showed that the fungal community colonising seedlings in container nurseries is dominated by Fusarium species.
Grey mould is an important pathogen, with polyphagous properties that allow it to infect a wide range of plant species, including forest plants, while its broad host range means it is found worldwide (Cheung et al., 2020). A study by Capieau et al. (2004) showed that grey mould is the most common economically important fungal disease in Swedish forest nurseries. For this reason, this species was classified as a primary pathogen in our study. Isolates of B. cinerea occurred on gnawed pine seedlings in virtually all study plots except the control sample, which may indicate that the seedlings are more susceptible to colonisation following tissue damage.
R. nigricans was another species that was regularly isolated from the material we analysed in our own studies. According to Lennartsson et al. (2014), fungi of the genus Rhizopous are considered “first colonisers” - a category of microorganisms that typically initially colonise easily accessible substrates such as decomposing and dead plant material. R. nigricans has hundreds of described hosts in dozens of plant families, including vegetables, ornamentals, fruits, bulbs, seeds, nuts and grains (Farr & Rossman, 2020).
Representatives of the genus Penicillium were also isolated from the pine seedlings gnawed by animals. Jankowiak (2008) suggested that Penicillium, Trichoderma and Mucor species as well as Hormonema dematioides Lagerb. & Melin could play an important role in the colonisation of pine tissues after an infestation with Tomicus piniperda (L.). In his investigations, he showed that there was significant colonisation of pine tissue with Penicillium fungi in the two weeks following the beetle infestation. The fact that the samples in our study were taken after the deer had fed, may have contributed to the lower proportion of Penicillium in the samples, which could explain the lower proportion of these fungi. In addition, Penicillium are an essential component of the natural microflora that colonise plant tissue and contribute to the biological defence of plants against phytopathogenic fungi due to their antagonistic properties (Guijarro et al., 2018; Win et al., 2021).
In our study, it was possible to detect the presence of Phytophthora spp. in pine seedlings gnawed by forest animals using the qPCR technique. This technique is very precise as it allows the detection of fg DNA quantities, which is equivalent to the detection of single fungal spores (Pollard & Okubara, 2019), and it is also effective in the detection of many plant disease-causing pathogens (Ji et al., 2022; Nowakowska et al., 2017; Okorski et al., 2022). Studies conducted under laboratory conditions have shown that various Phytophthora species have infectious capabilities against pine seedlings (Tkaczyk et al., 2016). A study by Chavarriaga et al. (2007) showed that P. cinnamomi can colonise natural pine stands. In our study, Phytophthora was found in a small number of samples, which could indicate an accidental introduction of infectious material to pine seedlings by forest animals.
Mycobiome differences with distance from agriculture
Plot 1, which was furthest away from an agricultural crop, was dominated by the following species: A. alternata, B. cinerea, F. avenaceum, R. nigricans and representatives of the genus Penicillium. The few species present in this study area were: Alternaria chartarum, Aureobasidium pullulans, Diplodia pinea, F. oxysporum, Lophodermium pini-mugonis, Pestalotia bicolor.
The mycoflora of the animal-infested pine seedlings in plot 3 (adjacent to the arable field where winter wheat was grown) was dominated by entophytic-saprotrophic species: C. cladosporioides, A. alternata and Penicillium sp. The group of endophytes as secondary pathogens on the seedlings from this plot was dominated by the species F. avenaceum. Only a few species were represented: Aureobasiudium bolleyi, Drechslera spp. F. equiseti, F. tricinctum and Nigrospora oryzae (Table 3). Analysis of the literature shows that the mycoflora of pine seedlings damaged by forest animals is similar to the fungi typical of winter wheat plants grown in Poland (Martyniuk et al., 2009, 2011). Based on this study, the group of authors confirmed the presence of species on wheat ears in both studies: A. alternata, A. tenuissima, B. cinerea, Cladosporium herbarum, F. poae, F. tricinctum and Penicillium sp. A similar composition of the mycoflora of winter wheat grains from north-east Poland was confirmed by Okorski et al. (2022) and Pszczółkowska et al. (2013).
In the case of plot 7, which also bordered an agricultural field where winter oilseed rape was grown, the mycoflora of the gnawed seedlings was dominated by endophytic fungi: F. avenaceum, A. alternata, B. cinerea, F. oxysporum and representatives of the genus Penicillium. Fungi that occurred in small numbers on cuttings from this plot were: Periconia macrospinosa, Diplodia pinea, Aspergillus niger and Alternaria tenuissima, which were classified as both saprotrophic endophytes and primary pathogens. The plot was directly adjacent to a winter oilseed rape crop. Studies by Mankeviciene et al. (2011) show that oilseed rape seeds are infected by Fusarium fungi, including F. avenaceum, F. culmorum, F. oxysporum, while B. cinnerea is a known polyphagous pathogen causing a widespread disease of oilseed rape in Poland - grey mould (Kurowski et al., 2010).
Based on the mathematical analysis, a trend was found suggesting that the infection level of gnawed pine seedlings was higher in the vicinity of agricultural crops, which could be related to the direct transmission of the inoculum by deer. However, proximity to agricultural crops, including winter wheat, may also have created favourable conditions for the direct transmission of spores to the seedlings, e.g. via air currents. We believe that cervids may play a role as vectors of plant pathogenic fungi, but in the absence of definitive evidence, research should continue to definitively address this question.
Fusarium avenaceum was most abundant in plots 7 and 9, which were closest to agricultural land (7 was adjacent to arable land, while 9 was about 200 m from agricultural fields). However, based on the mycological analysis, it is not possible to say directly how the pine seedlings were inoculated. It is likely that the transmission of the spores was the result of feeding, most probably by deer. However, it could also be a spontaneous transmission of the spores by the wind (Manstretta et al., 2015).
Colonisation and plant age
Our study found a negative correlation between the age of the seedlings and the amount of Fusarium DNA detected by qPCR. This may be explained by the fact that younger seedlings are more susceptible to infection by Fusarium species under severe stress conditions, such as the biting stress, which may have caused the healthy microbiome of the pine seedlings to transform into a troublesome pathobiont, which is important in forestry.
Advantages and disadvantages of the methods used for mycobiome analysis
This study used both cultural and molecular diagnostic methods to identify microorganisms in the forest seedlings. In our opinion, this was the right approach for this study, because these methods are complementary, as demonstrated in this study by performing linear correlation analyses between the amount of Fusarium fungal DNA and the number of cultures of these fungi isolated from pine seedlings.
It should be emphasised, however, that classical mycological diagnosis based on microscopic identification is very difficult. It is primarily based on culture morphology, colony colour, mycelial features, sporulation and conidial features on PDA medium including spore size or shape, which are not always unambiguous. Indeed, the morphological variability of individual strains within a species has led to the same species being repeatedly discovered by different researchers (Fang et al., 2020). On the other hand, morphological similarity of fungi can have the opposite effect - in the case of the genus Alternaria, for example, there is debate as to whether A. alternata and A. tenuissima are different species (Ozkilinc et al., 2017). It is also important to consider that environmental factors can influence traits such as spore length and width (Manawasinghe et al., 2021). Within the fungal genera, there are species that have many strong morphological features that allow easy differentiation, while on the other hand, there are species for which there are few diagnostic features and which are only available to experts specialised in genus identification (Ozkilinc et al., 2017). In recent years, there have been advances in the use of artificial intelligence to automatically identify the morphology of fungal hyphae from microscopic images (Koo et al., 2021).
The study of fungal diversity and taxonomy is fundamental to the development of science, but requires strong integration with the practical application of the results of such analyses (de Hoog et al., 2023). Studies on the phylogeny of fungi make it possible to organise systematics and gain insights into the adaptive trends of individual species. In recent years, the number of newly discovered fungal species described in the Index Fungorum has exceeded 3000 per year thanks to the use of sophisticated molecular biology methods such as NGS sequencing or nanopore sequencing (Phukhamsakda et al., 2022), and research in this field has helped to organise the systematics of important fungal genera such as Alternaria or Cladosporium (Htun et al., 2022; Sandoval-Denis et al., 2016). In the work of Aime et al. (2021) and Chethana et al. (2021), who proposed a detailed basis for the differentiation of new fungal species, a polyphasic approach was recommended that includes the combination of morphology, phylogeny, ecology, chemistry and other useful evidence.
Hyde et al. (2024) point out that numerous fungal taxa have been described based on only a few isolates, ignoring intraspecific variation. Studies based on only a single strain, incorrect species names or inadequate taxon selection are labelled as inappropriate approaches for distinguishing new taxa (Yasanthika et al., 2023). Therefore, when introducing a new species, it is recommended to use more than one strain in studies (Manawasinghe et al., 2021). It is estimated that up to 90% of fungal species diversity in different ecosystems has not yet been recognised and described. Therefore, there is an urgent need to sample as many previously undescribed species as possible to reconstruct the tree of fungal life (Zhou & May, 2022).
The qPCR method was applied to ascertain the presence or absence of certain Fusarium species and Phytophthora spp., which are of great importance due to their frequent occurrence on biting material in agricultural crops. The qPCR method is currently one of the most sensitive research techniques for analysing infectious material, which is used not only in agricultural phytopathology (Okorski et al., 2022), forestry (Nowakowska et al., 2017), but is also used in the detection of allergen-related food contamination (García et al., 2017) or in the investigation of animal (Yang et al., 2022) and human diseases (Biava et al., 2018). The disadvantage of the method is that the genetic material of dead microbial organisms is also amplified. It is therefore unclear at what stage the infection was at the time of sampling (Schena et al., 2013). To mitigate this, classical mycological tests were also carried out, as was the use of TaqMan qPCR diagnostics, being a research tool characterised by high sensitivity and accuracy.
In the case of Fusarium species, while infection was confirmed through both PCR analysis and the culture method, which could partially eliminate the above problem since only live organisms would be culturable, the question is not definitively resolved, as Fusarium can occur on plants as an endophyte (Zakaria & Ning, 2013) and in the case of forest trees as a secondary pathogen or even as a saprotroph (Yu et al., 2021).
Colonisation of forest plantations by agricultural pathogens and resulting problems
When seedlings planted in forest plantations die (presumably due to pathogenic fungi introduced from agricultural crops and also from infected soil used in nurseries), this causes measurable economic losses (Długosiewicz et al., 2019). According to the Forest Protection Manual (Leśne & Państwowe, 2012), in the event of losses of 20% or more, foresters must replenish the plantation with new seedlings, which is costly and time-consuming (Duryea & Landis, 2012). If the game pressure is high (e.g. because the hunting clubs do not cover their shooting requirements), foresters often have to fence such reforestation areas completely. Fencing is very costly and, although it completely excludes the forest animals from visiting the reforestation area, it also restricts their food source. However, fencing is necessary if the objective is to convert conifer monocultures into mixed forests, as the chances of successful establishment of deciduous tree species are minimal without this special, expensive protection.
In our study, endophytic fungi that can change their mode of life to pathogenic F. avenaceum, F. culmorum, F. equiseti, F. oxysporum and F. tricinctum were found on seedlings damaged by forest animals, which could indicate the invasion of these endophyte secondary pathogens from the winter wheat crop into the pine seedlings weakened by browsing.
Consequences of browsing and expected resistance reaction of seedlings to fungi
We assume that in the area where the observation plots were set up, roe deer in particular cause damage to crops, both in agriculture and in forest plantations. The population there is very dense and they feed in the fields at night and in the dense forest plantations during the day. Under these circumstances, their habitat is mainly restricted by human activities (fences, hunting) (Fuller & Gill, 2001). It has been estimated that up to 70% of damage to young trees is due to browsing (Ward et al., 2008). In forest plantations, the frequency of damage to conifer shoots by deer is positively correlated with animal density, and negatively correlated with the relative abundance of vegetation in the understorey (Ward et al., 2008). However, it was not the aim of this study to determine which deer species could be the disease vector, but we can speculate that roe deer are very likely due to their high population density in Poland, similar to other European countries. Konôpka and Pajtík (2015) reported that all the trees they examined showed browsing symptoms in the form of shedding of leading shoots or lateral branches. In our study, we chose most of the observation plots near agricultural land used by roe deer, which, unlike other deer, are territorial. One symptom of deer foraging was the shearing of pine shoots. It is estimated that almost all tree and shrub species in Poland and Europe are eaten by red deer (Budny et al., 2022; Pfeffer et al., 2022), while roe deer have a narrower food preference (Borkowski et al., 2021).
Various protective measures are taken to minimise game damage, such as the construction of wire mesh fences, permanent fencing, the establishment of game browsing buffer zones, but also the protection of the uppermost buds by various methods (Budny et al., 2022). The reduction of browsing by fences is usually applied to crops with small areas, as it is a costly procedure that also limits the availability of food for wild animals. In our trial, we did not use this method and the wild animals had free access to our observation plots. This allowed us to compare the mycobiome in plots located at different distances from the agricultural crops. In the study by Palmer and Truscott (2003), the authors showed that deer are the main cause of damage to pine seedlings in summer, with browsing symptoms occurring at a height of 1 m. At the same time, the researchers found that seedlings with earlier browsing were more likely to be broken than seedlings that had not been browsed before. We took the samples for the study in early autumn, which can undoubtedly affect the qualitative and quantitative composition of the fungal communities colonising the tissue of animal-damaged pine seedlings. The study by Baranowska et al. (2019) confirms that the fungal communities studied in autumn are characterized by a high abundance of fungal taxa. The consequences of browsing by cervids on the tree species they utilise are complex and not fully understood, including fungal infections (Gordon & Prins, 2008). When the leading shoot is browsed too heavily, the increased stress prolongs the time for normal growth, and in extreme cases this damage can open the door to infection with pathogenic fungi (Keča et al., 2018). While colonisation of damaged tissue may be a matter of chance (e.g. the occurrence of a wound in a particular place and time and the presence of pathogen spores), the consequences of infection can be quite different. Primary pathogens that can attack living tissue, e.g. Fusarium oxysporum on oak buds (Gorzkowska et al., 2016; Wit et al., 2015) produce mycotoxins that facilitate the killing and colonisation of tissue. However, they cannot decompose wood. Therefore, they constantly need new, living tissue to continue growing. They are followed by fungi that do not have the above-mentioned abilities but can break down lignin or cellulose or both. The browsing of seedlings in forest plantations is a gateway for pathogenic fungi. Most of them are opportunists and take advantage of the opportunity, and the destruction of the cover tissue undoubtedly facilitates the colonisation of the plant tissue. The trees are better able to protect themselves from gnawing or mechanical damage. This creates a zone of compartmentalisation of decomposition in the trees, which Shigo termed CODIT (Shigo, 1984). He claimed that when trees are damaged, they respond to the infested wood with chemical and physical changes to limit decomposition, which he termed compartmentalisation. The trees attempt to surround, isolate and contain the invader with phenolic compounds, which is the secret of tree longevity.
Multi-species interactions
Our research is part of a new approach to plant disease research that departs from the “one pathogen - one disease” paradigm, as identification of the pathogen is often insufficient to explain the syndromic state of multiple diseases (Pitlik & Koren, 2017; Vayssier-Taussat et al., 2015). This concept presents plants as holobionts. In a state of equilibrium, plant health is determined by the high diversity of microorganisms and the complex and highly intricate relationships between a plant and the microbiome that colonises its tissues. According to Bass et al. (2019), benign exogenous microorganisms can become key pathogenic organisms for the occurrence of diseases in opportunistic situations, so-called pathobionts. In our study, the biting stress may have caused the healthy microbiome of the pine seedlings to transform into a troublesome pathobiont. Fungi of the genus Fusarium, which were termed endophytes -secondary pathogens in this study, have the potential and ability to change their lifestyle from endophytic to pathogenic and cause disease due to their extensive pathogenic capabilities. Such a possibility is confirmed by the study of Okorski et al. (2015) on beech seedlings (Fagus sylvatica L.) in forest container nurseries. In it, they confirmed the selection of a highly virulent F. avenaceum genotype for beech, which caused a germination of up to 31% per cassette and a mortality of 5–7% of the seedlings. This was due to the introduction of horticultural plants into the forest nursery or the selection of the genotype due to the use of fungicides and the large scale of cultivation (5–10 million beech seedlings per year).
Conclusions
-
Pine seedlings under wild browsing stress can be colonised by fungi that are important pathogens of cultivated plants, such as Fusarium avenaceum, F. culmorum, F. graminearum and F. tricinctum.
-
Secondary pathogen fungal communities had higher values for the biodiversity indicators the closer the experimental plots were to agricultural land.
-
The importance of the primary pathogens of pine on gnawed tissue increased with increasing distance from the fields, with a positive correlation between relative abundance and dominance.
-
Younger pine seedlings that are gnawed are more heavily infested with Fusarium species than older seedlings, and also show lower species diversity and a dominance of certain secondary pathogen species.
-
Injuries to seedlings in plantations are a gateway for fungal infections that can more easily colonise exposed tissues.
-
The proximity between forestry and agricultural ecosystems can allow pathogens, particularly of the genus Fusarium, to colonise forest seedlings.
Data Availability
Details are available on request from the Department of Entomology, Phytopathology and Molecular Diagnostics, Faculty of Agriculture and Forestry University of Warmia and Mazury in Olsztyn Plac Łódzki 510-727 Olsztyn, Poland.
References
Aime, M. C., Miller, A. N., Aoki, T., Bensch, K., Cai, L., Crous, P. W., et al. (2021). How to publish a new fungal species, or name, version 3.0. IMA Fungus, 12, 1–15.
Arhipova, N., Jansons, A., Zaluma, A., Gaitnieks, T., & Vasaitis, R. (2015). Bark stripping of Pinus contorta caused by moose and deer: Wounding patterns, discoloration of wood, and associated fungi. Canadian Journal of Forest Research, 45, 1434–1438. https://doi.org/10.1139/cjfr-2015-0119
Baranowska, M., Korzeniewicz, R., Kartawik, N., & Behnke-Borowczyk, J. (2019). Seasons and fungal communities - beetles colonising stumps of American black cherry. Sylvan, 163(10), 872–880. https://doi.org/10.26202/sylvan.2019094
Bass, D., Stentiford, G. D., Wang, H. C., Koskella, B., & Tyler, C. R. (2019). The pathobiome in animal and plant diseases. Trends in Ecology & Evolution, 34, 996–1008. https://doi.org/10.1016/j.tree.2019.07.012
Biava, M., Colavita, F., Marzorati, A., Russo, D., Pirola, D., Cocci, A., Petrocelli, A., Guanti, M. D., Cataldi, G., Kamara, T., et al. (2018). Evaluation of a rapid and sensitive RT-qPCR assay for the detection of Ebola virus. Journal of Virological Methods, 252, 70–74. https://doi.org/10.1016/j.jviromet.2017.11.009
Bocianowski, J., Szulc, P., Waśkiewicz, A., & Cyplik, A. (2020). The effect of agrotechnical factors on Fusarium mycotoxins level in maize. Agriculture, 10, 528. https://doi.org/10.3390/agriculture10110528
Borkowski, J., Banul, R., Jurkiewicz-Azab, J., Hołdyński, C., Święczkowska, J., Nasiadko, M., & Załuski, D. (2021). There is only one winner: The negative impact of red deer density on roe deer numbers and distribution in the Słowiński National Park and its vicinity. Ecology and Evolution, 11, 6889–6899. https://doi.org/10.1002/ece3.7538
Budny, M., Zalewski, K., Skubis, J., Czaplicki, S., & Okorski, A. (2022). The content of long- chain hydrocarbons in the bark of selected willow cultivars in the context of cervids’ food preferences. Sylwan, 166, 309–320. https://doi.org/10.26202/sylwan.2022028
Buśko, M., Kulik, T., Ostrowska, A., Góral, T., & Perkowski, J. (2014). Quantitative volatile compound profiles in fungal cultures of three different Fusarium graminearum chemotypes. FEMS Microbiology Letters, 359, 85–93. https://doi.org/10.1111/15746968.12569
Capieau, K., Stenlid, J., & Stenström, E. (2004). Potential for biological control of Botrytis cinerea in Pinus sylvestris seedlings. Scandinavian Journal of Forest Research, 19(4), 312–319. https://doi.org/10.1080/02827580310019293
Chavarriaga, D., Bodles, W. J. A., Leifert, C., Belbahri, L., & Woodward, S. (2007). Phytophthora cinnamomi and other fine root pathogens in north temperate pine forests. FEMS Microbiology Letters, 276(1), 67–74.
Chethana, K. W. T., Manawasinghe, I. S., Hurdeal, V. G., & Bhunjun, C. S. (2021). What are fungal species and how to delineate them? Fungal Diversity, 109, 1–25.
Cheung, N., Tian, L., Liu, X., & Li, X. (2020). The destructive fungal pathogen Botrytis cinerea-insights from genes studied with mutant analysis. Pathogens, 9(11), 923. https://doi.org/10.3390/pathogens9110923
Czarnecka, D., Czubacka, A., Agacka-Mołdoch, M., Trojak-Goluch, A., & Księżak, J. (2022). The occurrence of fungal diseases in maize in organic farming versus an integrated management system. Agronomy, 12, 558. https://doi.org/10.3390/agronomy12030558
Davydenko, K., Nowakowska, J., Kaluski, T., Gawlak, M., Sadowska, K., García, J., Diez, J., Okorski, A., & Oszako, T. (2018). A comparative study of the pathogenicity of Fusarium circinatum and other Fusarium species in polish provenances of P. Sylvestris L. Forests, 9, 560. https://doi.org/10.3390/f9090560
de Hoog, S., Walsh, T. J., Ahmed, S. A., Alastruey-Izquierdo, A., Alexander, B. D., Arendrup, M. C., Babady, E., et al. (2023). A conceptual framework for nomenclatural stability and validity of medically important fungi: a proposed global consensus guideline for fungal name changes supported by ABP, ASM, CLSI, ECMM, ESCMID-EFISG, EUCAST-AFST, FDLC, IDSA, ISHAM, MMSA, and MSGERC. Journal of Clinical Microbiology, 61(11), e0087323. https://doi.org/10.1128/jcm.00873-23
Długosiewicz, J., Zając, S., & Wysocka-Fijorek, E. (2019). Ekonomiczna efektywność naturalnego i sztucznego odnowienia drzewostanów sosnowych w Nadleśnictwie Nowa Dęba. Sylwan, 163(05), 373–384.
Drenkhan, R., Ganley, B., Martín-García, J., Vahalík, P., Adamson, K., Adamčíková, K., Ahumada, R., Blank, L., Bragança, H., Capretti, P., et al. (2020). Global geographic distribution and host range of fusarium circinatum, the causal agent of pine pitch canker. Forests, 11, 724.
Duryea, M. L., & Landis, T. D. (Eds.). (2012). Forest nursery manual: production of bareroot seedlings (Vol. 11). Springer Science & Business Media.
Ellis, M. B. (1971). Dematiaceous hyphomycetes. Commonwealth Mycological Institute.
Elvira-Recuenco, M., Cacciola, S. O., Sanz-Ros, A. V., Garbelotto, M., Aguayo, J., Solla, A., Mullett, M., Drenkhan, T., Oskay, F., Aday Kaya, A. G., et al. (2019). Potential interactions between invasive fusarium circinatum and other pine pathogens in europe. Forests, 11, 7.
Fang, D. Z., Liu, X. L., Chen, X. R., Yan, W. W., He, Y. L., Cheng, Y., Chen, J., Li, Z. M., Guo, L. T., Wang, T. H., et al. (2020). Fusarium species and Fusarium oxysporum species complex genotypes associated with yam wilt in south-Central China. Frontiers in Microbiology, 11, 1964.
Farr, D.F., Rossman, A.Y., 2020. Fungal Databases, U.S. National Fungus Collections, ARS, USDA. Retrieved March 27, 2020, from https://nt.ars-grin.gov/fungaldatabases/. Accessed 13 May 2024.
Fuller, R. J., & Gill, R. M. (2001). Ecological impacts of increasing numbers of deer in British woodland. Forestry, 74, 193–199. https://doi.org/10.1093/forestry/74.3.193
García, A., Madrid, R., García, T., Martín, R., & González, I. (2017). Detection of food allergens by Taqman real-time PCR methodology. Food Allergens: Methods and Protocols, 95–108. https://doi.org/10.1007/978-1-4939-6925-8_8
Gerlach, W., Nirenberg, H., Eckart, I., Rummland, I., & Schwarz, R. (1982). In K. P. Parey (Ed.), The genus Fusarium: A pictorial atlas (Vol. 209).
Gordon, I. J., & Prins, H. H. (2008). The ecology of browsing and grazing. In Ecological Studies (Vol. 195). Springer. https://doi.org/10.1007/978-3-540-72422-3
Gorzkowska, A., Pszczółkowska, A., Okorski, A., Nowakowska, J. A., & Oszako, T. (2016). Fungi colonising oak seedlings (Quercus robur L.) in forest plantations in North-Eastern Poland. Folia Forestalia Polonica, 58, 79–86. https://doi.org/10.1515/ffp-2016-0009
Guijarro, B., Larena, I., Casals, C., Teixidó, N., Melgarejo, P., & Cal, A. D. (2018). Compatibility interactions between the biocontrol agent Penicillium frequentans Pf909 and other existing strategies to Brown rot control. Biological Control, 129, 45–54. https://doi.org/10.1016/j.biocontrol.2018.11.011
GUS, 2023. Production of agricultural and horticultural crops in 2022. Warszawa 2023.
Harrington, C.A., 1977. G77–380 growing conifers from seed. Historical Materials from University of Nebraska-Lincoln Extension 851.
Htun, A. A., Liu, H. F., He, L., et al. (2022). New species and new record of Alternaria from onion leaf blight in Myanmar. Mycological Progress, 21, 59–69. https://doi.org/10.1007/s11557-021-01765-x
Hyde, K. D., Baldrian, P., Yanpeng, C. Y., Thilini Chethana, K. W., Sybren De Hoog, S., et al. (2024). Current trends, limitations and future research in the fungi? Fungal Diversity, 125, 1–71. https://doi.org/10.1007/s13225-023-00532-5
Jankowiak, R. (2008). Fungi associated with Tomicus minor on Pinus sylvestris in Poland and their succession into the sapwood of beetle-infested windblown trees. Canadian Journal of Forest Research, 38(10), 2579–2588. https://doi.org/10.1139/x08-101
Ji, Y., Li, X., Gao, Q. H., et al. (2022). Colletotrichum species pathogenic to strawberry: Discovery history, global diversity, prevalence in China, and the host range of top two species. Phytopathology Research, 4, 42. https://doi.org/10.1186/s42483-022-00147-9
Keča, N., Tkaczyk, M., Żółciak, A., Stocki, M., Kalaji, H. M., Nowakowska, J. A., & Oszako, T. (2018). Survival of European ash seedlings treated with phosphite after infection with the Hymenoscyphus fraxineus and phytophthora species. Forests, 9, 442. https://doi.org/10.3390/f9080442
Konôpka, B., & Pajtík, J. (2015). Why was browsing by red deer more frequent but represented less consumed mass in young maple than in ash trees?! Journal of Forest Science, 61, 431–438. https://doi.org/10.17221/70/2015-jfs
Koo, T., Kim, M. H., & Jue, M. S. (2021). Automated detection of superfcial fungal infections from microscopic images through a regional convolutional neural network. PLoS One, 16, e0256290. https://doi.org/10.1371/journal.pone.0256290
Kulik, T. (2008). Detection of Fusarium tricinctum from cereal grain using PCR assay. Journal of Applied Genetics, 49(3), 305–311. https://doi.org/10.1007/BF03195628
Kulik, T., Jestoi, M., & Okorski, A. (2011). Development of TaqMan assays for the quantitative detection of fusarium avenaceum/fusarium tricinctum and fusarium poae esyn1 genotypes from cereal grain. FEMS Microbiology Letters, 314, 49–56. https://doi.org/10.1111/j.1574-6968.2010.02145.x
Kurowski, T. P., Majchrzak, B., & Jankowski, K. (2010). Effect of sulfur fertilization on the sanitary state of plants of the family Brassicaceae. Acta Agrobotanica, 63(1), 171–178.
Lee, K.-Y. (2023). Special issue: Relationship between insect vectors and plant pathogens. Archives of Insect Biochemistry and Physiology, 112, e21999. https://doi.org/10.1002/arch.21999
Lennartsson, P. R., Taherzadeh, M. J., & Edebo, L. (2014). Rhizopus. Encyclopedia of Food Microbiology, 284–290. https://doi.org/10.1016/b978-0-12-384730-0.00391-8
Leslie, J. F., & Summerell, B. A. (Eds.). (2006). The Fusarium laboratory manual. Blackwell Publishing. https://doi.org/10.1002/9780470278376
Leśne, P. G., & Państwowe, L. (2012). Instrukcja ochrony lasu (Forest Protection Manual). Lasy Polskie–2012.
Livak, K. J., & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- δδct method. Methods, 25, 402–408. https://doi.org/10.1006/meth.2001.1262
Malewski, T., Brzezińska, B., Belbahri, L., & Oszako, T. (2019). Role of avian vectors in the spread of Phytophthora species in Poland. European Journal of Plant Pathology, 155(4), 1363–1366.
Manawasinghe, I. S., Phillips, A. J., Xu, J., & Balasuriya, A. (2021). Defining a species in fungal plant pathology: Beyond the species level. Fungal Diversity, 109, 267–282.
Mancini, V., & Romanazzi, G. (2014). Seed treatments to control seedborne fungal pathogens of vegetable crops. Pest Management Science, 70, 860–868.
Mankeviciene, A., Suproniene, S., Brazauskiene, I., & Gruzdeviene, E. (2011). Natural occurrence of Fusarium mycotoxins in oil crop seed. Plant Breeding and Seed Science, 63, 109–116.
Manstretta, V., Gourdain, E., & Rossi, V. (2015). Deposition patterns of Fusarium graminearum ascospores and conidia within a wheat canopy. European Journal of Plant Pathology, 143, 873–880. https://doi.org/10.1007/s10658015-0722-8
Martyniuk, S., Oroń, J., & Kozieł, M. (2011). Fungi, including Fusarium spp., on ears of conventionally and ecologically grown winter wheat. Plant Breeding and Seed Science, 63(−1). https://doi.org/10.2478/v10129-011-0023-0
Martyniuk, S., Oroń, J., & Mączka, M. (2009). Populations of fungi and bacteria on ears of conventionally and organically grown winter wheat. Phytopathologia, 51, 53–61.
Neupane, S., Andersson, B., Högberg, N., Ihrmark, K., & Alström, S. (2013). Fungal communities associated with field grown oilseed rape (Brassica napus L.) - their possible role in early crop establishment. Acta Agriculturae Scandinavica, Section B - Soil & Plant Science, 63, 241–252. https://doi.org/10.1080/09064710.2012.756117
Nowakowska, J., Malewski, T., Tereba, A., & Oszako, T. (2017). Rapid diagnosis of pathogenic Phytophthora species in soil by real-time PCR. Forest Pathology, 47, e12303. https://doi.org/10.1111/efp.12303
Okorski, A., Milewska, A., Pszczółkowska, A., Karpiesiuk, K., Kozera, W., Dąbrowska, J. A., & Radwińska, J. (2022). Prevalence of Fusarium fungi and Deoxynivalenol levels in winter wheat grain in different climatic regions of Poland. Toxins, 14, –102. https://doi.org/10.3390/toxins14020102
Okorski, A., Polak-Śliwińska, M., Karpiesiuk, K., Pszczółkowska, A., & Kozera, W. (2017). Real time PCR: A good tool to estimate mycotoxin contamination in pig diets. World Mycotoxin Journal, 10, 228–219. https://doi.org/10.3920/wmj2016.2137
Okorski, A., Pszczółkowska, A., Gorzkowska, A., Okorska, S., & Głuszek, P. (2019). Fungi associated with conifer seedlings grown in forest nurseries under different systems. Environmental Engineering & Management Journal (EEMJ), 18, 1509–1517.
Okorski, A., Pszczółkowska, A., Okorska, S., & Fordoński, G. (2015). First report of Fagus sylvatica infection by Fusarium avenaceum in Forest container nurseries in northeastern Poland. Plant Disease, 99(3), 420. https://doi.org/10.1094/PDIS-09-14-0899-PDN
Olaizola, J., Pajares, J. A., Gordon, T. R., & Diez, J. J. (2023). Aggressiveness of Fusarium oxysporum and F. Verticillioides isolates on stone and scots pine under greenhouse conditions. Forest Pathology, 53, e12831. https://doi.org/10.1111/efp.12831
Ozkilinc, H., Rotondo, F., Pryor, B. M., & Peever, T. L. (2017). Contrasting species boundaries between sections Alternaria and Porri of the genus Alternaria. Plant Pathology, 67(2), 303–314. https://doi.org/10.1111/ppa.12749
Palmer, S., & Truscott, A. M. (2003). Browsing by deer on naturally regenerating scots pine (Pinus sylvestris L.) and its effects on sapling growth. Forest Ecology and Management, 182, 31–47. https://doi.org/10.1016/S0378-1127(03)00026-4
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Research, 29, e45–e45. https://doi.org/10.1093/nar/29.9.e45
Pfeffer, S. E., Dressel, S., Wallgren, M., Bergquist, J., & Kalén, C. (2022). Browsing damage on scots pine: Direct and indirect effects of landscape characteristics, moose and deer populations. Diversity, 14, 734. https://doi.org/10.3390/d14090734
Pfeffer, S. E., Singh, N. J., Cromsigt, J. P., Kalén, C., & Widemo, F. (2021). Predictors of browsing damage on commercial forests–a study linking nationwide management data. Forest Ecology and Management, 479, 118597. https://doi.org/10.1016/j.foreco.2020.118597
Phukhamsakda, C., Nilsson, R. H., Bhunjun, C. S., de Farias, A. R. G., Sun, Y. R., Wijesinghe, S. N., Raza, M., et al. (2022). The numbers of fungi: Contributions from traditional taxonomic studies and challenges of metabarcoding. Fungal Diversity, 114, 327–386.
Pitlik, S. D., & Koren, O. (2017). How holobionts get sick—Toward a unifying scheme of disease. Microbiome, 5, 1–4. https://doi.org/10.1186/s40168-017-0281-7
Pollard, A. T., & Okubara, P. A. (2019). Real-time PCR quantification of Fusarium avenaceum in soil and seeds. Journal of Microbiological Methods, 157, 21–30. https://doi.org/10.1016/j.mimet.2018.12.009
Pszczółkowska, A., Okorski, A., Olszewski, J., & Jarmołkowicz, J. (2013). Fungal pathogens of the genus Fusarium in winter wheat protected with fungicides in North-Eastern Poland. Acta Agrobotanica, 66(2), 95–106.
Ramos, J. A., Bugalho, M. N., Cortez, P., & Iason, G. R. (2006). Selection of trees for rubbing by red and roe deer in forest plantations. Forest Ecology and Management, 222, 39–45.
Reid, A. (2006). Sampling and testing for plant pathogens. Department of Primary Industries and Regional Development, Western Australia, Perth. Bulletin, 4683.
Roques, A., Fan, J. T., Courtial, B., Zhang, Y. Z., Yart, A., Auger-Rozenberg, M. A., et al. (2015). Planting sentinel European trees in eastern Asia as a novel method to identify potential insect pest invaders. PLoS One, 10(5), e0120864.
Sandoval-Denis, M., Gené, J., Sutton, D. A., Wiederhold, N. P., Cano-Lira, J. F., & Guarro, J. (2016). New species of Cladosporium associated with human and animal infections. Persoonia, 36, 281–298. https://doi.org/10.3767/003158516X691951
Scarlett, K., Tesoriero, L., Daniel, R., & Guest, D. (2014). Sciarid and shore flies as aerial vectors of Fusarium oxysporum f. sp. cucumerinum in greenhouse cucumbers. Journal of Applied Entomology, 138, 368–377. https://doi.org/10.1111/jen.12098
Schena, L., Li Destri Nicosia, M., Sanzani, S., Faedda, R., Ippolito, A., & Cacciola, S. (2013). Development of quantitative PCR detection methods for phytopathogenic fungi and oomycetes. Journal of Plant Pathology, 7–24.
Shigo, A. L. (1984). Compartmentalization: A conceptual framework for understanding how trees grow and defend themselves. Annual Review of Phytopathology, 22, 189–214. https://doi.org/10.1146/annurev.py.22.090184.001201
Sikora, K., Verstappen, E., Mendes, O., Schoen, C., Ristaino, J., & Bonants, P. (2012). A universal microarray detection method for identification of multiple Phytophthora spp. using padlock probes. Phytopathology, 102, 635–645. https://doi.org/10.1094/PHYTO-11-11-0309
Šnepsts, G., Krišāns, O., Matisons, R., Seipulis, A., & Jansons, A. (2022). Cervid bark-stripping is an explicit amplifier of storm legacy effects in Norway spruce (Picea abies (L.) karst.) stands. Forests, 13. https://doi.org/10.3390/f13111947
Statistics Poland. (2021). Statistical yearbook of forestry. Statistical Office in Bialystok, Podlaskie Centre for Regional Surveys https://stat.gov.pl. (accessed on 15 August 2022)
Tkaczyk, M., Sikora, K., Nowakowska, J. A., Aniśko, E., Oszako, T., Belbahri, L., & Milenković, I. (2016). Four different Phytophthora species that are able to infect scots pine seedlings in laboratory conditions. Folia Forestalia Polonica Series A – Forestry, 58(3), 123–130.
Vayssier-Taussat, M., Kazimirova, M., Hubalek, Z., Hornok, S., Farkas, R., Cosson, J. F., Bonnet, S., Vourch, G., Gasqui, P., Mihalca, A. D., et al. (2015). Emerging horizons for tick-borne pathogens: From the ‘one pathogen–one disease’vision to the pathobiome paradigm. Future Microbiology, 10, 2033–2043. https://doi.org/10.2217/fmb.15.114
Vegi, A., & Wolf-Hall, C. E. (2013). Multiplex real-time PCR method for detection and quantification of mycotoxigenic fungi belonging to three different genera. Journal of Food Science, 78, M70–M76. https://doi.org/10.1111/j.17503841.2012.03008.x
Ward, A. I., White, P. C., Walker, N. J., & Critchley, C. H. (2008). Conifer leader browsing by roe deer in English upland forests: Effects of deer density and understorey vegetation. Forest Ecology and Management, 256, 1333–1338. https://doi.org/10.1016/j.foreco.2008.06.034
Watanabe, T. (2010). Pictorial atlas of soil and seed fungi: Morphologies of cultured fungi and key to species. CRC press.
Win, T. T., Bo, B., Malec, P., & Fu, P. (2021). The effect of a consortium of Penicillium sp. and Bacillus spp. in suppressing banana fungal diseases caused by Fusarium sp. and Alternaria sp. Journal of Applied Microbiology, 131, 1890–1908.
Wit, M., Sierota, Z., Oszako, T., Mirzwa-Mróz, E., Wakuliński, W., et al. (2015). Fusarium spp. on the above-ground organs of dying oaks-a new threat? Sylwan, 159, 403–410.
Yamoah, E., Jones, E. E., Suckling, D. M., Bourdôt, G. W., Walter, M., & Stewart, A. (2011). Using insects as potential vectors of Fusarium tumidum to control gorse. New Zealand Entomologist, 34(1), 5–11. https://doi.org/10.1080/00779962.2011.9722202
Yang, L., Long, Y., Li, Q., Song, W., Huang, Y., Liang, G., Yu, D., Zhou, M., Xu, G., Chen, Y., et al. (2022). Detection of suid herpesvirus 1 infectivity in pigs by propidium monoazide-qPCR. Frontiers in Veterinary Science, 9. https://doi.org/10.3389/fvets.2022.975726
Yasanthika, W. E., Tennakoon, D. S., De Farias, A. R. G., & Chethana, K. T. (2023). Isolation and characterization of novel Dothideomycetes species from forest soils in Chiang Rai and Krabi (Thailand): Additions to the diversity of Curvularia and Verruconis. Phytotaxa, 609, 17–44.
Yu, Z., Ding, H., Shen, K., Bu, F., Newcombe, G., & Liu, H. (2021). Foliar endophytes in trees varying greatly in age. European Journal of Plant Pathology, 160, 375–384. https://doi.org/10.1007/s10658-021-02250-7
Zakaria, L., & Ning, C. H. (2013). Endophytic Fusarium spp. from roots of lawn grass (Axonopus compressus). Tropical Life Sciences Research, 24, 85.
Zhang, S., Zheng, Q., Xu, B., & Liu, J. (2019). Identification of the fungal pathogens of postharvest disease on peach fruits and the control mechanisms of bacillus subtilis jk-14. Toxins, 11, 322.
Zhou, L. W., & May, T. W. (2022). Fungal taxonomy: Current status and research agendas for the interdisciplinary and globalisation era. Mycology, 14(1), 52–59. https://doi.org/10.1080/21501203.2022.2103194
Funding
The results presented in this paper were obtained as part of a comprehensive study financed by the University of Warmia and Mazury in Olsztyn (grant No. 30.610.009.110).
Author information
Authors and Affiliations
Contributions
Conceptualization, A.O.,T.O and G.P.; methodology, A.O.,T.O and G.P.; software, P.B.,A.R.; validation, P.B.,A.R.A.P.; formal analysis, A.O.,J.D.,G.P. investigation, A.O.,J.D.,G.P.; resources, A.R.; data curation, P.B.,A.R.; writing—original draft preparation, A.O.,T.O. P.B.; writing—review and editing, A.O.,T.O.; visualization, P.B.,A.R.; supervision, A.O.,T.O.; project administration, A.O.; funding acquisition, A.P., T.O. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A
Appendix A
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Okorski, A., Paczkowska, G., Dąbrowska, J. et al. Characterization of the mycobiome of Pinus sylvestris L. seedlings damaged by forest animals, with an emphasis on pathogenic species. Eur J Plant Pathol (2024). https://doi.org/10.1007/s10658-024-02940-y
Accepted:
Published:
DOI: https://doi.org/10.1007/s10658-024-02940-y