Abstract
Centrosome amplification (CA), an abnormal increase in the number of centrosomes in the cell, is a recurrent phenomenon in lung and other malignancies. Although CA promotes tumor development and progression by inducing genomic instability (GIN), it also induces mitotic stress that jeopardizes cellular integrity. CA leads to the formation of multipolar mitotic spindles that can cause lethal chromosome segregation errors. To sustain the benefits of CA by mitigating its consequences, malignant cells are dependent on adaptive mechanisms that represent therapeutic vulnerabilities. We aimed to discover genetic dependencies associated with CA in lung cancer. Combining a CRISPR/Cas9 functional genomics screen with tumor genomic analyses, we identified the motor protein KIFC1, also known as HSET, as a putative vulnerability specifically in lung adenocarcinoma (LUAD) with CA. KIFC1 expression was positively correlated with CA in LUAD and associated with worse patient outcomes, smoking history, and indicators of GIN. KIFC1 loss-of-function sensitized LUAD cells with high basal KIFC1 expression to potentiation of CA, which was associated with a diminished ability to cluster extra centrosomes into pseudo-bipolar mitotic spindles. Our work suggests that KIFC1 inhibition represents a novel approach for potentiating GIN to lethal levels in LUAD with CA by forcing cells to divide with multipolar spindles, rationalizing further studies to investigate its therapeutic potential.
Similar content being viewed by others
Introduction
Lung cancer (LC) is the leading cause of cancer death worldwide [1] and is broadly divided into small-cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). Of the various NSCLC subtypes, lung adenocarcinoma (LUAD) is most prevalent, accounting for ~40% of all cases [2]. Although advances in targeted therapies and immunotherapy have expanded the treatment landscape for LC, the 5-year net survival rate has not risen above 22% [3]. This highlights the need for new therapeutic approaches that target mechanisms supporting lung tumor growth and progression.
Centrosomes are microtubule-organizing organelles with important roles in cell motility and polarity, and generation of mitotic spindles required to segregate chromosomes during mitosis [4]. Centrosome amplification (CA), an abnormal increase in the number of centrosomes in a cell, is a common feature of many malignancies including LC [4,5,6]. Several routes to CA have been described including DNA damage, failure of cytokinesis, centriole overduplication, and carcinogen exposure [7,8,9,10]. The presence of extra centrosomes has been shown to promote genomic instability (GIN) [11, 12] which fuels tumor adaptation to stress such as that induced by anti-cancer therapy [13]. This fitness advantage may explain the prevalence of CA in cancer; however, having supernumerary centrosomes comes with a cost. Cells with CA are prone to forming multipolar mitotic spindles that if left uncorrected cause severe chromosome segregation errors, mitotic catastrophe, and cell death [14]. Thus, cancer cells must manage extra centrosomes to prevent multipolar divisions and their deleterious effects in order to sustain its tumor-promoting effects. Several coping mechanisms have been proposed with clustering of extra centrosomes into pseudo-bipolar spindles being the most well established [4]. As such, the proteins regulating these adaptive mechanisms represent vulnerabilities and potential therapeutic targets in cancer cells with CA.
Multiple reports have demonstrated the occurrence and clinical relevance of CA in LC [5, 6]. However, despite its occurrence, the mechanisms LC use to cope with CA have not been directly investigated. We sought to identify collateral dependencies associated with the presence of extra centrosomes in LC models. Integrating a CRISPR/Cas9 screen with tumor genomic analyses, we identified KIFC1 (also known as HSET) as a putative survival factor for LUAD with CA. We discovered that KIFC1 inactivation is lethal specifically in LC cells with high basal KIFC1 expression. Our analyses of multiple cell lines and independent tumor datasets revealed that KIFC1 expression is positively correlated with CA in LUAD suggesting centrosome number could also indicate LC sensitivity to KIFC1 inhibition. KIFC1 depletion sensitized LUAD cells with high endogenous KIFC1 expression to potentiation of CA and this phenotype was associated with a diminished ability to cluster extra centrosomes. Collectively, our findings nominate KIFC1 as a therapeutic target in LUAD with high basal KIFC1 levels, rationalizing the development of KIFC1-specific inhibitors and further preclinical investigation of its therapeutic potential.
Materials And Methods
Tumor and cell line genomic analyses
RNA-sequencing (RNASeqV2) and associated clinical data for The Cancer Genome Atlas (TCGA) Pan-Cancer (PANCAN) cohort were accessed from the National Cancer Institute’s Genomic Data Commons portal [15]. Fraction of genome altered (FGA), aneuploidy scores (AS), and mutation status for the most common oncogenic drivers (EGFR, KRAS, MET, ROS1, RET, ALK, NRAS and BRAF) in TCGA LUAD tumors were obtained from cBioPortal [16]. Gene expression and essentiality data for the Cancer Cell Line Encyclopedia (CCLE) were obtained from the DepMap database (https://depmap.org/portal/) [17]. Briefly, DepMap gene effect scores were calculated from CRISPR screen data using the Chronos algorithm. More negative gene effect scores indicate a greater dependency on the gene. Normalized microarray-derived gene expression data for 4 additional LUAD cohorts were obtained from the Gene Expression Omnibus (GSE50081, GSE31210, GSE68465, GSE75037). CA20 gene expression signature scores for TCGA lung tumors and CCLE cell lines were obtained from a published study [18], and were calculated for additional cohorts as the sum of normalized, median-centered expression levels for the 20 signature genes (AURKA, CCNA2, CCND1, CCNE2, CDK1, CEP63, CEP152, E2F1, E2F2, LMO4, MDM2, MYCN, NDRG1, NEK2, PIN1, PLK1, PLK4, SASS6, STIL and TUBG1), as described [19].
Immunohistochemistry
Tumor tissues were banked after informed written patient consent following a protocol approved by the University Health Network Research Ethics Board. Formalin-fixed paraffin embedded tissues from 10 NSCLC patient-derived xenograft (PDX) models were processed for immunohistochemistry to detect centrosomes using the anti-pericentrin antibody ab4448 (Abcam, 1:1000 dilution). Mitotic cells were classified as having CA if they contained more than 2 centrosomes and the proportion with CA was calculated for each tumor. At least 30 mitotic cells were counted per tumor tissue section.
Cell culture and lentivirus production
Cell lines were grown at 37 °C and 5% CO2 in DMEM, Hams F12 or RPMI 1640 media supplemented with 10% fetal bovine serum, 2 mM glutamine, and 100 U/ml of penicillin-streptomycin. NCI-H1299, H1975, H1944, H2291, H1792, H2122, H23, H2030, H1568, H1650, HCC827, HCC2935 were generous gifts from the late Dr. Adi Gazdar. PC9 was gifted from Dr. William Lockwood and A549 was gifted from Dr. Haibo Zhang. Cell line identities were confirmed using short tandem repeat profiling. Cells were confirmed to be negative for mycoplasma using a PCR-based assay [20]. Platinum-E (PE) cells were obtained from Cell BioLabs and used to produce lentivirus by transfection with psPAX2 and pMD2.G (Addgene #12260, #12259) using Lipofectamine 3000.
CRISPR/Cas9 genetic screen
H1299 cells were engineered with stable Cas9 expression using Lenti-Cas9-blast (Addgene #52962). Efficient nuclease activity was confirmed using a reporter system (Addgene #67979, #67980; Supp Fig. 1). We synthesized a custom single guide RNA (sgRNA) knockout (KO) library with 13,243 sgRNA targeting 3319 genes that are “druggable” with FDA-approved or investigational compounds annotated in the DrugBank and bindingDB databases [21, 22], as well as negative controls (sgLacZ, sgLuciferase, sgGFP) (Supplementary Table 1). This library was cloned into pRC0162 (Addgene #195319) and sequence verified. H1299-Cas9 cells were infected with the library at a multiplicity of infection of 30% for single integrations per cell to yield 500X library coverage. Following 48 h of puromycin selection, cells were pooled and split into 3 replicates each for treatment with vehicle (DMSO) or 7.5 nM of CFI-400945 (945) [23]. Ten million cells were collected at time zero (T0) and the remainder were cultured in the presence of DMSO or 945 for 17 cell doublings and then harvested for targeted sgRNA sequencing. Screen hits were identified using the DrugZ algorithm [24].
Competition assays
H1975, H2030, H2122, PC9, and A549 were engineered to express functional Cas9 as above. sgRNA targeting AAVS1 (AV, negative control) or KIFC1 (K3, K4) were cloned into LentiGuide-puro-NLS-GFP or LentiGuide-puro-NLS-mCherry (#185473, 185474) and transduced into Cas9+ cells. sgRNA sequences are provided in Supplementary Table 2. mCherry+ AV (wildtype, WT) were mixed 1:1 with GFP + K3/K4/AV cells to compare the relative fitness of cells with and without KIFC1 perturbation. Cells were treated with DMSO or 7.5 nM 945 and mCherry/GFP+ ratios were measured by flow cytometry over multiple passages.
KIFC1 knockdown, dose response, and viability assays
Human Silencer Select siRNA assays s7906, s7907, and Negative Control No. 1 (Thermo Fisher Scientific) were used to knockdown (KD) KIFC1. Cells were transfected with siRNA using Lipofectamine RNAiMAX (50 nM in H1299, H1975, H2030, A549, H2122; 75 nM in PC9) and 24 h later, were sampled to confirm KD and plated for functional experiments. Dose response, clonogenic survival, and AZ82 cytotoxicity assays were done using sulforhodamine B staining as previously described [25]. Proliferation was assessed using the ATP-Lite assay.
KIFC1 overexpression
The KIFC1 coding sequence was PCR amplified from H1299 cDNA and TOPO cloned into pCR8/GW/TOPO (Invitrogen, #K250020). Primer sequences are listed in Supplementary Table 2. A Gateway LR Clonase recombination reaction (Invitrogen, #11791020) was performed to insert the coding sequence of KIFC1 or LacZ into pInducer20-Neomycin (Addgene #44012). Lentivirus was made as described and stable lines were selected and maintained in RPMI media prepared with tetracycline-free FBS containing 0.2 mg/ml neomycin.
Immunofluorescence, immunoblotting, and real-time quantitative PCR
Immunofluorescence (IF) was used to detect centrosomes using the following antibodies: CEP192 (1:1000, Bethyl Labs#A302-324A), ɑ-tubulin (1:1000, Sigma-Aldrich #T9026), goat anti-mouse AlexaFluor-488, and goat anti-rabbit AlexaFluor-555. For IF to assess KIFC1 localization, centrosomes were detected by γ-tubulin (1:1000, Sigma-Aldrich #T5326), microtubules with ɑ-tubulin (1:1000, SantaCruz SC-53029), and KIFC1 with Abcam AB172620 (1:200 dilution) and goat anti-mouse AlexaFluor-488, goat anti-rat AlexaFluor-647 and goat anti-rabbit AF555. DNA was stained with DAPI. CA was calculated as described for IHC above. Western blots were done using the following antibodies: KIFC1 (1:1000, CST#12313), GAPDH (1:5000, CST#97166), and β-ACTIN (1:5000, Proteintech#66009-1-IG). RT-qPCR to confirm siRNA-mediated KIFC1 KD was done using HPRT1 as the endogenous control. Primer sequences are listed in Supplementary Table 2.
Statistics
Statistical tests and the number of replicates for each experiment are described in the figure legends. All statistics and plots were generated using R and GraphPad Prism software. For cell culture assays, 2-6 independent experimental replicates, each with 2-6 technical replicates, were done to ensure adequate power and reproducibility of results. T-tests were done on continuous data from independent experimental replicates assumed to be random representative samples from a normally distributed dataset with equal variance. All t-tests were unpaired and two-tailed. Chi-square tests were done on categorical data, with the categories being mutually exclusive, with all data points representing a random sampling of independent observations. Mann Whitney U tests were performed with two-tails on continuous data assumed to be non-normally distributed and representing a randomly drawn sampling from a population with equal variance. Kruskal-Wallis tests were done to compare continuous data across 3 or more groups with the assumption that data were randomly sampled, groups were mutually exclusive, and data were not normally distributed. Survival analyses were done using nonparametric log-rank (Mantel-Cox) tests with the assumption that survival times in each group are independent. Correlation analyses were done on continuous data using Pearson’s method with the assumptions that both variables were normally distributed and linearly related. An ANOVA was performed with KIFC1 expression as the continuous response variable, age as a continuous variable, and driver gene status, sex, tumor stage, and smoking history as categorical factors. A cox proportional hazards model was used to estimate associations between survival time and several clinical factors (sex, stage, smoking history, driver mutation status), and KIFC1 expression and age as continuous variables. For CRISPR screen analyses, significant hits were identified as genes having a DrugZ FDR q-value < 0.25. For all other statistical tests, p < 0.05 was considered significant with *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. All methods were performed in accordance with the relevant guidelines and regulations.
Results
Centrosome amplification (CA) is prevalent in NSCLC tissues
Previous studies have reported centrosome abnormalities in 29–53% of NSCLC tissues [5, 6]. We evaluated CA in 10 patient-derived xenograft (PDX) models of NSCLC (4 LUAD, 3 squamous cell carcinoma (LUSC), and 3 large cell carcinoma) using IHC to detect the established centrosome marker, pericentrin [26, 27]. Cases were chosen to represent different NSCLC subtypes. Our assessment of these tumors confirmed that CA is prevalent in NSCLC (Fig. 1A). CA was detected in mitotic cells from all 10 PDX, with the percentage of cells with CA ranging from 18 to 66% (Fig. 1B). We observed that centrosomes in cells with CA were disorganized in multipolar spindles or clustered into pseudo-bipolar spindles (Fig. 1A). The presence of CA in clinical NSCLC tissues rationalized investigation of vulnerabilities associated with extra centrosomes in cell models.
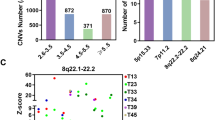
A Representative IHC images of mitotic cells in NSCLC tissues from patient-derived xenografts (PDX) stained for the centrosome marker pericentrin. Arrowheads point to centrosomes. Scale bar indicates 10 um. B Characterization of CA in 10 NSCLC PDX. Centrosomes were scored in mitotic cells only and cells were classified as having CA if they contained > 2 pericentrin foci. C Potentiation of CA in H1299-Cas9+ cells by 945 treatment during the CRISPR screen. Cells were sampled from each replicate (N = 3/treatment arm). Bar plots indicate mean with SD, asterisks indicate t-test p < 0.0001. D Representative IF images exhibiting bipolar (BP), multipolar (MP), and pseudo-bipolar (P-BP) mitotic spindles in H1299 cells. Scale bar indicates 5 um. E Characterization of mitotic spindles in H1299-Cas9+ cells sampled during the CRISPR screen. Counts were pooled across the 3 replicates for each treatment condition. Asterisks indicate chi-square test p < 0.0001. A minimum of 160 total mitotic cells were counted for each treatment condition for IF analyses in (C, E). F Normalized Z-scores for each gene targeted in the sgRNA library calculated with the DrugZ algorithm. Z-scores represent the relative abundance of sgRNAs in cells treated with 945 relative to DMSO control. Negative Z-scores indicate genes targeted by sgRNAs that were significantly depleted in cells in which CA was potentiated by 945. G Correlations between gene expression and CA20 scores for the top 16 genes identified by the CRISPR screen in the TCGA LUAD cohort. Correlation coefficients indicate those for Pearson’s correlation analyses.
CRISPR screen identifies KIFC1 as a putative CA-coping factor in LC
To discover genetic dependencies in LC cells with CA, we performed a CRISPR/Cas9 screen in the LUAD cell line, H1299. We selected H1299 because of its high Cas9 editing efficiency (Supp Fig. 1A), its relatively high basal CA which implied adaptation to CA (Fig. 1C; Supplementary Fig. 4C), and its amenability to pharmacological potentiation of CA (Supp Fig. 1B). We used low dose CFI-400945 (945), a PLK4 inhibitor, to potentiate CA to levels seen in clinical tumor tissues (Fig. 1A, B), as cell lines are known to exhibit less CA [28] (Supp Fig. 1B). H1299-Cas9 cells were infected with a custom sgRNA KO library targeting ~3300 “druggable” genes, grown in the presence of DMSO or sublethal dose of 945 (7.5 nM) that potentiated CA for 17 cell doublings (Supp Fig. 1C), and then harvested for sgRNA sequencing. We first compared sgRNA abundance in DMSO-treated cells at end point to cells harvested at screen onset (T0) to confirm the depletion of sgRNA targeting essential genes. This identified MYC as the top hit with 7 other genes classified by the DepMap as “common essential genes” ranking in the top 10 hits (CDC25, SRSF1, RPL4, PSMA3, RPL14, ATP2A2, and SCD) [17] (Supp Fig. 1D, E). Notably, NRAS, an established oncogenic driver in H1299 [17] was identified as the 10th ranked hit (Supp Fig. 1D, E). Detection of these essential genes confirmed the technical success of our CRISPR/Cas9 screen.
Sampling of H1299 cells during the screen confirmed that 7.5 nM 945 potentiated CA to ~65% of cells and increased the incidence of multipolar spindles compared to DMSO (Fig. 1C, E). Accordingly, growth in the presence of 945 was slower (Supplementary Fig. 1C). We reasoned that cells containing sgRNA targeting genes essential for survival in the presence of extra centrosomes would be selectively depleted in populations treated with 945 compared to DMSO. Analysis of sgRNA abundance in 945- versus DMSO-treated populations identified 16 putative CA survival genes with an FDR q-value < 0.25 (Fig. 1F; Supplementary Table 3). To prioritize hits from our screen, we examined their correlations with a gene expression signature of CA called CA20 in the TCGA LUAD cohort [29]. CA20 is a composite of 20 genes associated with the presence of CA that was validated in clinical breast tumors [18, 19]. We hypothesized that genes important for coping with extra centrosomes would be highly expressed in tumors with CA and positively correlated with CA20 scores. Of the 16 candidates, none were CA20 signature genes and KIFC1 exhibited the strongest positive correlation with CA20 in LUAD (Fig. 1G; Supplementay Fig. 2A). KIFC1 is a minus-end directed kinesin motor protein involved in mitotic spindle assembly and organization, as well as vesicle and organelle trafficking [30]. It also has a well-described role in centrosome clustering in cancer cells [30]. Thus, our CA sensitization screen suggested that H1299 cells are dependent on KIFC1 to cope with CA, nominating KIFC1 as a therapeutic vulnerability in LC with supernumerary centrosomes.
KIFC1 is associated with clinical factors in LUAD
We next aimed to determine the clinical relevance of KIFC1 in patient-derived tumor tissues by analyzing KIFC1 mRNA expression in the TCGA LUAD cohort. Consistent with previous reports in pan-cancer or LC-focused studies [31,32,33], KIFC1 was significantly overexpressed in LUAD compared to non-malignant lung tissues (Fig. 2A), and LUAD patients whose tumors had high KIFC1 expression had significantly worse 5 year survival outcomes than those with low expression in a univariate model (Fig. 2B). KIFC1 expression remained significantly associated with overall survival in a Cox-proportional hazards model including additional prognostic factors including age, stage, sex and smoking history (Table 1). Expression was also elevated in tumors of greater stage, and for the first time, we discovered that expression was significantly higher in smokers relative to non-smokers (Fig. 2C, D). Lastly, we found that KIFC1 expression was modestly elevated in males relative to females and in tumors without oncogenic driver mutations compared to those with oncogenic alterations (Supplementary Fig. 2B, C). An ANOVA suggested that smoking history was the clinical factor most strongly associated with KIFC1 expression (Table 2). Although KIFC1 was overexpressed and correlated with CA20 in LUSC, we did not observe any significant associations between KIFC1 expression and patient characteristics or survival (Supp Fig. 3). We then investigated the robustness of these associations in 4 independent LUAD cohorts with gene expression and clinical data available (GSE75037, GSE50081, GSE31210, GSE68465) (Supp Fig. 2D–H). These analyses confirmed that high KIFC1 expression was associated with smoking (3/4 cohorts), advanced stage (4/4 cohorts), and worse 5-year survival (4/4 cohorts). Together these findings indicate the clinical and prognostic significance of KIFC1 in LUAD.
KIFC1 expression was analyzed in the TCGA LUAD cohort to investigate associations with malignancy (A), 5-year survival (B), stage (C), and smoking history (D). Asterisks indicate significance for Mann Whitney or Kruskal-Wallis tests (**p < 0.01, ****p < 0.0001). Error bars indicate mean and SD. NS Never smokers, CS Current smokers, FS < 15 Former smoker for fewer than 15 years, FS > 15 Former smoker for over 15 years. The association between KIFC1 expression and outcome was assessed in patients whose tumors exhibited KIFC1 expression ranking in the top and bottom tertiles of expression. Asterisks indicate p < 0.01 for a log-rank test on the Kaplan-Meier survival curves.
CA and GIN correlate with KIFC1 expression in LUAD models and tissues
Since our screen nominated KIFC1 as a genetic dependency of LC cells with CA, we suspected KIFC1 expression would be higher in cells with extra centrosomes. To test this hypothesis, we evaluated KIFC1 protein expression and CA in a panel of LUAD cell lines. Both KIFC1 expression and CA were variable across models (Supplementary Fig. 4A–C). In support of our hypothesis, we identified a positive correlation between KIFC1 protein expression and CA across 14 LUAD cell lines (Fig. 3A). We next evaluated the association between CA20 and KIFC1 dependency scores from the DepMap database in a larger panel of LC cell lines from the Cancer Cell Line Encyclopedia (CCLE) collection [17]. Our analyses revealed no association between CA20 and KIFC1 essentiality across 52 NSCLC or 13 LUSC lines, but indicated trends towards a greater dependency on KIFC1 in 15 LUAD and 11 small-cell lung cancer cell lines predicted to have high levels of CA (Fig. 3B, C; Supplementary Fig 4D, E). Thus, KIFC1 may be a LUAD- and SCLC-specific dependency. As noted above, we observed a strong positive correlation between KIFC1 expression and CA20 in LUAD tumors from the TCGA (Fig. 1G; Supplementary Fig. 2A), and we validated this correlation in 4 independent cohorts (Supplementary Fig. 2G). Because CA drives genomic instability (GIN) [11, 12], we also investigated associations between KIFC1 expression and measures of GIN in the TCGA LUAD cohort. This revealed significant positive correlations between KIFC1 and fraction of genome altered (FGA) and aneuploidy scores (Fig. 3D, E). Collectively, these findings demonstrate that KIFC1 expression is associated with CA and GIN in LUAD models and tissues.
A Correlation between KIFC1 expression and CA in 14 LUAD models. Protein expression was quantified from western blots and CA from IF conducted under basal conditions (Supplementary Fig. 4). B, C Correlation between KIFC1 dependency and CA20 in NSCLC (B) and LUAD (C) cell lines from the CCLE. KIFC1 gene effect scores were accessed from the DepMap database and negative scores indicate genetic dependency. CA20 scores were obtained from the de Almeida 2019 study. D, E Correlation between KIFC1 mRNA expression and fraction of genome altered (FGA) and aneuploidy score. FGA and aneuploidy scores were obtained from cBioPortal. All correlation coefficients indicated were calculated using Pearson’s correlation analyses.
LUAD cells are dependent on KIFC1 for survival upon potentiation of CA
We next sought to functionally validate KIFC1 as a dependency of LC cells with CA using in vitro growth competition assays that mimic CRISPR screen conditions. We used models with different endogenous levels of KIFC1 and examined the effects of KIFC1 loss-of-function (LOF) on cellular fitness. H1299, H1975, and H2030 were selected as representative models with relatively high basal KIFC1 expression, and A549, PC9, and H2122 as models with low KIFC1 (Supplementary Fig. 4). Treatment with low-dose 945 potentiated CA and increased the proportion of multipolar spindles in each LUAD line except A549 (Fig. 4A, B). Cas9+ derivatives of these lines were transduced with a GFP+ lentivector encoding independent sgRNA targeting KIFC1 (K3 or K4), or a GFP+ or mCherry+ lentivector encoding an sgRNA targeting AAVS1 (AV, wildtype (WT) control). CRISPR-mediated KIFC1 inactivation was evident in all models relative to controls (Fig. 4C). We competed GFP + WT or GFP + K3/K4 cells with mCherry+ WT cells in the presence of DMSO or 945 and monitored the GFP:mCherry+ ratio over several passages. These experiments validated KIFC1 as a hit from our screen, as GFP+ cells were depleted in the 945-treatment condition over time (Fig. 4D), confirming that KIFC1 LOF sensitized H1299 cells to potentiation of CA. We observed a similar phenotype in H1975 and H2030 with high basal KIFC1 expression. However, KIFC1 perturbation did not reduce the fitness of PC9, H2122, or A549 cells with relatively low endogenous KIFC1 levels (Fig. 4D). These results implicate KIFC1 as a specific dependency when CA is potentiated in LUAD cell lines with high KIFC1 expression. The lack of sensitization in H2122 and PC9, despite modest increases in CA by 945, suggests these models may use KIFC1-independent mechanisms to cope with multipolar spindles caused by CA, while the lack of effect in A549 likely reflects their resistance to CA induction.
A Potentiation of CA by low-dose 945 treatment (7.5 nM) in Cas9+ cell models. Centrosomes were detected using IF for CEP192 and CA was scored as the proportion of cells with greater than two centrosomes. Error bars indicate the mean and SEM (N = 3 for H1299, H1975, PC9, H2122, A549; N = 2 for H2030), and asterisks indicate p-value significance for t-tests between treatments. B Classification of mitotic spindles as bipolar (BP), multipolar (MP), or pseudo-bipolar (P-BP) for each treatment condition. Asterisks indicate p-value significance for chi-square tests. For (A, B), data for the independent replicates are pooled and at least 100 total mitotic cells were scored for each treatment condition. C Confirmation of CRISPR-mediated loss of KIFC1 expression in LUAD cell lines. Cas9+ cells were transduced with mCherry (mCH) or GFP lentivectors to express sgRNA targeting AAVS1 (AV) or KIFC1 (K3 targeting exon 3 and K4 targeting exon 4). D Multicolour competition assays comparing the relative fitness of cells with wildtype (AV) and genetically perturbed KIFC1 (K3, K4) in DMSO- and 945-treated (7.5 nM) conditions. mCH-AV cells were mixed 1:1 with GFP-AV, GFP-K3, or GFP-K4 cells and grown over multiple passages. The GFP+ ratio was monitored over time by flow cytometry. Data are representative of two independent experiments for each model (*p < 0.05, ***p < 0.001, ****p < 0.0001).
Depletion of KIFC1 impairs cell viability and suppresses centrosome clustering in LUAD cell lines with potentiation of CA
Although multiple compounds have been reported to inhibit KIFC1 [34], their weak potencies require treating cells at high doses that preclude attributing treatment-induced phenotypes specifically to KIFC1 inhibition [35]. We investigated the effects of AZ82, the KIFC1 inhibitor reported to have the best specificity [35], on LUAD viability. We observed cytotoxic effects of AZ82 at 7.5uM but they did not correlate with KIFC1 expression (Supplementary Fig. 5). For example, PC9 exhibited the greatest sensitivity to AZ82 in contrast to the results of our CRISPR screen validation experiments (Fig. 4D; Supplementary Fig. 5). As such, we used two independent siRNAs to knockdown (KD) KIFC1 to investigate its therapeutic potential. Both siRNA yielded KD in all 6 models (Fig. 5A; Supplementary Fig. 6A). Dose response curves indicated that KIFC1 depletion enhanced sensitivity to 945 in H1299 (Supplementary Fig. 6C). A similar albeit less pronounced effect was observed in H2030 and H1975. KIFC1 KD had no effect on the sensitivity of A549 to 945, and little effect on H2122 and PC9, both of which were relatively resistant to 945 (i.e. < 40% killing; Supplementary Fig. 6C). We also conducted clonogenic survival assays to measure the effects of KIFC1 KD on cell viability. KIFC1 KD significantly reduced colony survival in H1299, H1975, and H2030 when CA was potentiated by 945 treatment, but had no effect in 945-treated PC9, H2122, and A549 cells (Fig. 5B). In DMSO-treated conditions without potentiation of CA, siRNA-mediated depletion of KIFC1 had no effect on clonogenic survival or cell proliferation (Supplementary Fig. 6B, D).
A Confirmation of siRNA-mediated KIFC1 knockdown by western blotting. NTC = non-targeting control. K1, K2 indicate independent siRNA targeting KIFC1. B Clonogenic survival assays. Data are representative of 3 (H1299, H2030, H2122, A549), 4 (PC9), or 5 (H1975) biological replicates per model. Colony survival was normalized to that observed in DMSO-treated conditions for each siRNA. Error bars indicate SEM of 6 technical replicates. C Classification of mitotic spindles as bipolar (BP), multipolar (MP), or pseudo-bipolar (P-BP). Data are pooled for 3 (H2030, H1975, H2122, PC9, A549) or 4 (H1299) replicate experiments and at least 150 total mitotic cells were scored for each treatment condition. Asterisks indicate p-value significance for chi-square tests between NTC and K1 or K2 siRNA for each treatment condition (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
CA can lead to the formation of multipolar spindles during mitosis (Fig. 1D, E) and KIFC1 counters their deleterious effects by clustering extra centrosomes to promote viable chromosome segregation and cell division [4]. We suspected that the dependence of LUAD cells with CA on KIFC1 was due to its role in centrosome clustering, so we evaluated the effects of KIFC1 KD on spindle polarity (Fig. 5C). KIFC1 depletion in H1299 and H2030 led to a significant decrease in centrosome clustering evident by a reduced proportion of mitotic cells with pseudo-bipolar spindles and an increased proportion of cells with multipolar spindles; a similar trend was seen in H1975 (Fig. 5C). These effects were more pronounced when CA was potentiated with 945 treatment. KIFC1 KD did not affect centrosome clustering in PC9, H2122, or A549 cells treated with DMSO (Fig. 5C). In 945-treated cells, KD of KIFC1 in PC9 and A549 increased the proportion of multipolar spindles but this phenotype was not reproduced with both siRNAs and the fold-change in multipolar spindles was less than that observed for H1299, H1975 and H2030 (Fig. 5C). KIFC1 KD in H2122 cells treated with 945 had no effect on the proportion of cells with pseudo-bipolar or multipolar spindles (Fig. 5C). Finally, we engineered H1299 cells with dox-inducible KIFC1 expression and treated them with 945 to potentiate CA. This revealed that overexpression of KIFC1 significantly reduced the proportion of cells with multipolar spindles (Supplementary Fig. 7). Together, these data indicate that KIFC1 promotes survival of LUAD models with high KIFC1 expression when CA is potentiated to levels seen in clinical tumors by clustering extra centrosomes into pseudo-bipolar spindles to avoid the lethal consequences of cell divisions with multipolar spindles.
Discussion
Centrosome amplification (CA) is a recurrent feature of many malignancies that promotes genomic instability (GIN), and consequently, cancer development and progression [4]. Not surprisingly, CA is associated with poor prognosis in many cancer types [18, 36]. Despite the prevalence of CA in lung tumors [5, 6], the vulnerabilities imposed by CA in LC and its therapeutic potential have not been defined. By integrating a CRISPR/Cas9 screen, genomic analyses of LUAD tumors, and functional studies in multiple cell lines, we identified and validated KIFC1 as a dependency of LUAD with high KIFC1 expression and supernumerary centrosomes. Kinesin family member C1 (KIFC1 or HSET) belongs to the kinesin-14 family of minus-end directed motor proteins and facilitates organization of the mitotic spindle [37]. Although KIFC1 is not essential in non-malignant cells, it is indispensable in some cancer cell lines [38]. Studies of multipolar divisions in cells with CA have demonstrated that KIFC1 clusters extra centrosomes into spindles with pseudo-bipolar arrangements by “ratcheting” microtubules emanating from different centrosomes to group them together [38,39,40,41,42,43]. Briefly, KIFC1 localizes to the spindle poles during mitosis (Supplementary Fig. 8) where it contributes to both spindle morphogenesis and focusing through its microtubule crosslinking and minus-end directed motor activity [39, 40, 42]. Both the tail and motor domains of KIFC1 bind to microtubules, allowing it to crosslink microtubules [37]. In cells with CA, KIFC1’s crosslinking function mediates stable connections between extra centrosomes [41]. During spindle formation, Cep215 directly binds to KIFC1 and captures KIFC1-bound kinetochore fibres (k-fibers), anchoring centrosomes to the poles of the mitotic spindle. This anchoring boosts centrosome-spindle pole stability and contributes to clustering efficiency [43]. Following attachment of extra centrosomes to the spindle poles, KIFC1 arranges them into a pseudo-bipolar formation by ATP-hydrolysis driven, minus-end directed movement along crosslinked microtubules from different centrosomes [39]. KIFC1’s efficiency in maintaining centrosome-centrosome cohesion and spindle focusing is supported by its interactions with the intraflagellar transport proteins, IFT52 and IFT88, as depletion of IFT52 or IFT88, or inhibition of KIFC1, increases multipolar spindle formation [41]. Collectively, these findings suggest a mechanism whereby KIFC1’s microtubule crosslinking and motor functions establish stable centrosome-spindle pole and centrosome-centrosome attachments to facilitate centrosome clustering.
Our discovery of KIFC1 as a putative vulnerability in LC is consistent with reports in breast, prostate, and ovarian cancers [34]. Liu and colleagues investigated KIFC1 using functional studies in 3 NSCLC lines and reported that KIFC1 depletion reduced viability and induced G2/M arrest and p21 expression but they did not evaluate centrosomes [32]. However, they reported that KIFC1 KD reduced viability to the greatest extent in H1299 cells, which we found to have relatively high basal CA and KIFC1 expression among the LUAD models we assessed. Advancing on correlative studies that did not consider centrosome number, our unique CA-focused approach nominated KIFC1 as a dependency specifically when CA is potentiated in LUAD models with high KIFC1 expression, attributable to its role in mitigating CA-associated mitotic stress via centrosome clustering. Patel et al. identified KIFC1 as a putative vulnerability in breast cancers based on integrative genomic analyses of tissues and cell lines. Similar to our finding, this group showed that KIFC1 was preferentially essential in triple-negative breast cancer models with CA [44]. Although the CA20 gene expression signature is only a surrogate for CA, our assessment of KIFC1 dependency and CA20 in the DepMap database further suggested that KIFC1 is a CA-specific dependency in LUAD and possibly SCLC [17]. KIFC1’s classification as a strongly selective dependency but not a common essential gene in DepMap likely indicates that a small fraction of cancer models with high basal CA are dependent on KIFC1. Notably, the 6 lines we studied harbor different oncogenic driver mutations, namely NRAS (H1299), KRAS (H2030, H2122, A549), and EGFR (H1975, PC9). These mutations were split amongst KIFC1-dependent (H1299, H1975, H2030) and -independent (H2122, PC9, A549) models, suggesting that KIFC1-dependency is not associated with LUAD driver genes. However, functional studies with a larger number of models representing various oncogenic mutations are required to confirm this, as our sample size is too small to determine whether specific mutations influence KIFC1 dependency in LUAD.
Liu et al. also reported that KIFC1 was overexpressed in NSCLC compared to non-malignant lung tissues and that high expression was associated with worse prognosis, which is consistent with findings from other LC studies [32, 33, 45, 46] and reports in other malignancies [31, 33, 34]. Our analyses of multiple LUAD cohorts confirmed associations of KIFC1 with clinical factors including survival and stage as well as CA20. It is important to note that some of the associations reported, including those that we have identified, used RNA as a readout of KIFC1 expression, while others used protein levels measured by IHC. Multiple independent studies have shown that the prognostic association between high KIFC1 and poor survival in lung cancer patients is robust whether RNA or protein expression is evaluated [32, 33, 45, 46]. However, to our knowledge, the correlation between KIFC1 mRNA and protein expression in LUAD tissues has not been formally assessed. Thus, studies to verify whether KIFC1 mRNA levels reliably indicate protein expression are warranted, especially to guide potential future use of KIFC1 expression as a clinical biomarker. For the first time, we found that KIFC1 expression was elevated in LUAD from patients with a smoking history. This observation was reproducible across multiple cohorts, raising the hypothesis that tobacco smoke could increase KIFC1 expression, possibly as a consequence of inducing CA. Although cigarette-smoke carcinogens are known to induce CA [47], experiments to test their effects on KIFC1 expression are required to formally test this hypothesis. Celik et al. recently reported that KIFC1 was overexpressed in 59% of lung tumors and that 96% of these exhibited hypomethylation of KIFC1 [45]. This suggests that recurrent overexpression of KIFC1 in LC is driven by epigenetic mechanisms, but this remains to be validated in additional cohorts. An integrative pan-cancer study by Wu et al. found that KIFC1 was rarely perturbed at the genetic level in LUAD and other tumor types, suggesting genetic mechanisms are unlikely to account for KIFC1 overexpression [31]. Further studies are needed to decipher the mechanisms driving KIFC1 overexpression but ample evidence indicates that KIFC1 expression is a robust prognostic biomarker in LC.
Targeting kinesins that regulate spindle assembly in cancer cells is a promising therapeutic approach particularly in genomically unstable tumors [48,49,50,51]. Inhibitors of KIF18A and Eg5 (encoded by KIF11) are being investigated in clinical trials [49, 52]. Our work nominates KIFC1 as a therapeutic target in LC, with KIFC1 and/or CA as a putative predictive biomarker. Our finding that KIFC1 expression was significantly higher in LUAD lacking common driver mutations and in smokers compared to former and never smokers suggests that KIFC1-targeted therapy could be useful in these currently underserved patient populations. Encouragingly, KIFC1 KD does not affect proliferation or survival in non-malignant cells in vitro [38, 44], but studies of KIFC1-targeted therapy in animal models are required to confirm its therapeutic index. None of the KIFC1 inhibitors described to date (e.g. AZ82, CW069, SR31527, and KAA) have progressed to clinical studies, which may be attributable to poor potencies and selectivities [34]. Thus, development of clinical-grade KIFC1 inhibitors is warranted, especially given their potential utility in diverse cancers.
Our analyses of KIFC1 expression and CA suggest that various opportunities exist to use KIFC1 and CA as potential prognostic biomarkers or predictors of response to KIFC1-targeted therapy. As described above, tumor RNA or protein expression of KIFC1 may prove useful for biomarker purposes. CA could be assessed in tumors using the RNA-based CA20 gene signature [19] or by physically scoring CA [36]. To score CA in tumor tissues, most studies have used immunostaining for pericentrin, gamma-tubulin, or centrin, which are components of the pericentriolar material and centrioles of centrosomes [36]. Because centriole overduplication is a prominent mechanism that gives rise to CA, enumeration of centrioles has been proposed as a gold standard assay for quantifying CA [53]. However, a study by Singh et al. demonstrated that pericentrin is a robust marker for centrosomes and quantifying CA [27]. Since scoring CA in clinical tissues is technically simpler and amenable to automated analyses when pericentrin is used to detect centrosomes [27], pericentrin staining may be the ideal marker for developing an IHC-based assay to measure CA in tumors. Validation of KIFC1 expression or CA as biomarkers to predict tumor response to KIFC1-targeted therapy could facilitate its clinical translation by informing rational patient selection.
Our work and that of others indicates that cultured cancer cells exhibit less CA than malignant cells in tumor tissues [28]. Accordingly, we treated LC cells with low dose 945 [23] to potentiate CA to better mimic clinical tumors and observed that KIFC1 LOF sensitized cells to CA. This observation was consistent with a study that reported that combining KIFC1 KD with potentiation of CA by cisplatin enhanced cytotoxicity in TNBC cells [44]. Together, these results suggest that KIFC1 inhibition could synergize with chemotherapy and/or irradiation which potentiate CA and/or multipolar spindles [10, 54, 55]. Supporting this concept, a recent study showed that KIFC1 KD sensitized LC to radiotherapy, though CA was not measured [56]. Further studies to define rational and effective KIFC1 inhibitor combinations are warranted, but our work nominates 945 as a promising CA-inducing drug for combination with KIFC1-targeted therapy. Confirming whether 945 treatment enhances CA in tumor tissues from treated patients would provide additional evidence to support this strategy. Since non-malignant cells are relatively resistant to 945-induced CA this combination could be clinically tractable [23, 57].
The recurrence of CA and KIFC1 overexpression in lung tumors, and the dependence of LC with high KIFC1 expression on KIFC1 when CA is potentiated emphasize its therapeutic potential. We propose that KIFC1-targeted therapy could provide a novel strategy for potentiating GIN to lethal levels by forcing LC to divide with multipolar spindles, and that KIFC1 expression and CA could serve as biomarkers to guide effective use. We believe the efficacy of KIFC1-targeted therapy will be greatest when combined with treatments that exacerbate CA in lung tumors, and suggest that developing clinical-grade KIFC1 inhibitors and identifying and evaluating rational therapeutic combinations is warranted.
Data availability
Datasets analyzed are available from public sources as described, and CRISPR screen data are available from the corresponding author upon reasonable request.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49.
Politi K, Herbst RS. Lung cancer in the era of precision medicine. Clin Cancer Res. 2015;21:2213–20.
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33.
Sabat-Pośpiech D, Fabian-Kolpanowicz K, Prior IA, Coulson JM, Fielding AB. Targeting centrosome amplification, an Achilles’ heel of cancer. Biochem Soc Trans. 2019;47:1209–22.
Koutsami MK, Tsantoulis PK, Kouloukoussa M, Apostolopoulou K, Pateras IS, Spartinou Z, et al. Centrosome abnormalities are frequently observed in non-small-cell lung cancer and are associated with aneuploidy and cyclin E overexpression. J Pathol. 2006;209:512–21.
Jung CK, Jung JH, Lee KY, Kang CS, Kim M, Ko YH, et al. Centrosome abnormalities in non-small cell lung cancer: correlations with DNA aneuploidy and expression of cell cycle regulatory proteins. Pathol Res Pr. 2007;203:839–47.
Godinho SA, Pellman D. Causes and consequences of centrosome abnormalities in cancer. Philos Trans R Soc Lond B Biol Sci. 2014; 369. https://doi.org/10.1098/rstb.2013.0467.
Liao W-T, Lin P, Cheng T-S, Yu H-S, Chang LW. Arsenic promotes centrosome abnormalities and cell colony formation in p53 compromised human lung cells. Toxicol Appl Pharm. 2007;225:162–70.
Thaiparambil J, Dong L, Jasso D, Huang J-A, El-Zein RA. Mitotic Spindle Apparatus Abnormalities in Chronic Obstructive Pulmonary Disease Cells: A Potential Pathway to Lung Cancer. Cancer Prev. Res. 2020. https://doi.org/10.1158/1940-6207.CAPR-19-0557.
Löffler H, Fechter A, Liu FY, Poppelreuther S, Krämer A. DNA damage-induced centrosome amplification occurs via excessive formation of centriolar satellites. Oncogene. 2013;32:2963–72.
Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–82.
Lingle WL, Barrett SL, Negron VC, D’Assoro AB, Boeneman K, Liu W, et al. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci USA. 2002;99:1978–83.
Sansregret L, Vanhaesebroeck B, Swanton C. Determinants and clinical implications of chromosomal instability in cancer. Nat Rev Clin Oncol. 2018;15:139–50.
Holland AJ, Cleveland DW. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep. 2012;13:501–14.
Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang T-H, et al. The Immune Landscape of Cancer. Immunity. 2018;48:812–30.e14.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1.
Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a Cancer Dependency Map. Cell. 2017;170:564–76.e16.
de Almeida BP, Vieira AF, Paredes J, Bettencourt-Dias M, Barbosa-Morais NL. Pan-cancer association of a centrosome amplification gene expression signature with genomic alterations and clinical outcome. PLoS Comput Biol. 2019;15:e1006832.
Ogden A, Rida PCG, Aneja R. Prognostic value of CA20, a score based on centrosome amplification-associated genes, in breast tumors. Sci Rep. 2017;7:262.
Uphoff CC, Drexler HG. Detecting mycoplasma contamination in cell cultures by polymerase chain reaction. Methods Mol Biol. 2011;731:93–103.
Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–82.
Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:D198–201.
Mason JM, Lin DC-C, Wei X, Che Y, Yao Y, Kiarash R, et al. Functional characterization of CFI-400945, a Polo-like kinase 4 inhibitor, as a potential anticancer agent. Cancer Cell. 2014;26:163–76.
Colic M, Wang G, Zimmermann M, Mascall K, McLaughlin M, Bertolet L, et al. Identifying chemogenetic interactions from CRISPR screens with drugs. Genome Med. 2019;11:52.
Thu KL, Silvester J, Elliott MJ, Ba-Alawi W, Duncan MH, Elia AC, et al. Disruption of the anaphase-promoting complex confers resistance to TTK inhibitors in triple-negative breast cancer. Proc Natl Acad Sci USA. 2018;115:E1570–7.
Denu RA, Zasadil LM, Kanugh C, Laffin J, Weaver BA, Burkard ME. Centrosome amplification induces high grade features and is prognostic of worse outcomes in breast cancer. BMC Cancer. 2016;16:47.
Singh A, Denu RA, Wolfe SK, Sperger JM, Schehr J, Witkowsky T, et al. Centrosome amplification is a frequent event in circulating tumor cells from subjects with metastatic breast cancer. Mol Oncol. 2020;14:1898–909.
Mittal K, Choi DH, Ogden A, Donthamsetty S, Melton BD, Gupta MV, et al. Amplified centrosomes and mitotic index display poor concordance between patient tumors and cultured cancer cells. Sci Rep. 2017;7:43984.
Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014; 511: 543–50.
Xiao Y-X, Yang W-X. KIFC1: a promising chemotherapy target for cancer treatment? Oncotarget. 2016;7:48656–70.
Wu H, Duan Y, Gong S, Zhu Q, Liu X, Liu Z. An Integrative Pan-Cancer Analysis of Kinesin Family Member C1 (KIFC1) in Human Tumors. Biomedicines 2022;10. https://doi.org/10.3390/biomedicines10030637.
Liu Y, Zhan P, Zhou Z, Xing Z, Zhu S, Ma C, et al. The overexpression of KIFC1 was associated with the proliferation and prognosis of non-small cell lung cancer. J Thorac Dis. 2016;8:2911–23.
Sun Y, Zhang Y, Lang Z, Huang J, Zou Z. Prognostic and clinicopathological significance of kinesin family member C1 in various cancers: A meta-analysis. Medicine. 2019;98:e17346.
Sharma N, Setiawan D, Hamelberg D, Narayan R, Aneja R. Computational benchmarking of putative KIFC1 inhibitors. Med Res Rev. 2023;43:293–318.
Yukawa M, Yamauchi T, Kurisawa N, Ahmed S, Kimura K-I, Toda T. Fission yeast cells overproducing HSET/KIFC1 provides a useful tool for identification and evaluation of human kinesin-14 inhibitors. Fungal Genet Biol. 2018;116:33–41.
Mittal K, Kaur J, Jaczko M, Wei G, Toss MS, Rakha EA, et al. Centrosome amplification: a quantifiable cancer cell trait with prognostic value in solid malignancies. Cancer Metastasis Rev. 2020. https://doi.org/10.1007/s10555-020-09937-z.
She Z-Y, Yang W-X. Molecular mechanisms of kinesin-14 motors in spindle assembly and chromosome segregation. J Cell Sci. 2017;130:2097–110.
Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–203.
Goshima G, Nédélec F, Vale RD. Mechanisms for focusing mitotic spindle poles by minus end–directed motor proteins. J Cell Biol. 2005;171:229–40.
Cai S, Weaver LN, Ems-McClung SC, Walczak CE. Kinesin-14 family proteins HSET/XCTK2 control spindle length by cross-linking and sliding microtubules. Mol Biol Cell. 2009;20:1348–59.
Vitre B, Taulet N, Guesdon A, Douanier A, Dosdane A, Cisneros M, et al. IFT proteins interact with HSET to promote supernumerary centrosome clustering in mitosis. EMBO Rep. 2020;21:e49234.
Mountain V, Simerly C, Howard L, Ando A, Schatten G, Compton DA. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol. 1999;147:351–66.
Chavali PL, Chandrasekaran G, Barr AR, Tátrai P, Taylor C, Papachristou EK, et al. A CEP215-HSET complex links centrosomes with spindle poles and drives centrosome clustering in cancer. Nat Commun. 2016;7:11005.
Patel N, Weekes D, Drosopoulos K, Gazinska P, Noel E, Rashid M, et al. Integrated genomics and functional validation identifies malignant cell specific dependencies in triple negative breast cancer. Nat Commun. 2018;9:1044.
Celik B, Pasin O, Sen S, Tuncer SB, Kayım ZY, Erciyas SK, et al. DNA methylation of KIFC1 gene in determination of histological diagnosis, prognosis and metastasis of lung cancer. Pathol Res Pr. 2023;249:154742.
Tabassum G, Singh P, Gurung R, Hakami MA, Alkhorayef N, Alsaiari AA, et al. Investigating the role of Kinesin family in lung adenocarcinoma via integrated bioinformatics approach. Sci Rep. 2023;13:9859.
Shinmura K, Iwaizumi M, Igarashi H, Nagura K, Yamada H, Suzuki M, et al. Induction of centrosome amplification and chromosome instability in p53-deficient lung cancer cells exposed to benzo[a]pyrene diol epoxide (B[a]PDE). J Pathol. 2008;216:365–74.
Tischer J, Gergely F. Anti-mitotic therapies in cancer. J Cell Biol. 2019;218:10–11.
Shahin R, Aljamal S. Kinesin spindle protein inhibitors in cancer: from high throughput screening to novel therapeutic strategies. Future Sci OA. 2022;8:FSO778.
Cohen-Sharir Y, McFarland JM, Abdusamad M, Marquis C, Bernhard SV, Kazachkova M, et al. Aneuploidy renders cancer cells vulnerable to mitotic checkpoint inhibition. Nature. 2021;590:486–91.
Payton M, Belmontes B, Hanestad K, Moriguchi J, Chen K, McCarter JD, et al. Small-molecule inhibition of kinesin KIF18A reveals a mitotic vulnerability enriched in chromosomally unstable cancers. Nat Cancer 2023. https://doi.org/10.1038/s43018-023-00699-5.
Braun J, Möckel MM, Strittmatter T, Marx A, Groth U, Mayer TU. Synthesis and biological evaluation of optimized inhibitors of the mitotic kinesin Kif18A. ACS Chem Biol. 2015;10:554–60.
Denu RA, Shabbir M, Nihal M, Singh CK, Longley BJ, Burkard ME, et al. Centriole Overduplication is the Predominant Mechanism Leading to Centrosome Amplification in Melanoma. Mol Cancer Res. 2018;16:517–27.
Mullee LI, Morrison CG. Centrosomes in the DNA damage response—the hub outside the centre. Chromosome Res. 2016;24:35–51.
Scribano CM, Wan J, Esbona K, Tucker JB, Lasek A, Zhou AS, et al. Chromosomal instability sensitizes patient breast tumors to multipolar divisions induced by paclitaxel. Sci Transl Med. 2021;13:eabd4811.
Wang X, Raman N, Lemtiri-Chlieh G, Chang J, Jagtap S, Chowdhury DD, et al. Griseofulvin Radiosensitizes Non-Small Cell Lung Cancer Cells and Activates cGAS. Mol Cancer Ther. 2023;22:519–28.
Kawakami M, Mustachio LM, Zheng L, Chen Y, Rodriguez-Canales J, Mino B, et al. Polo-like kinase 4 inhibition produces polyploidy and apoptotic death of lung cancers. Proc Natl Acad Sci USA. 2018;115:1913–8.
Acknowledgements
We thank the UHN Therapeutics group for providing CFI-400945 for our studies, TCGA for tumor genomics data, Dr. Xiaofeng Lu from UHT Research Core Facilities for assistance with IF, and the late Dr. Adi Gazdar for sharing LUAD cell lines.
Funding
This work was supported by funds from St. Michael’s Hospital, the Canada Research Chairs program, the Canadian Foundation for Innovation, and the Canadian Institutes of Health Research (CIHR). CZ and SSKB were supported by scholarships from the Research Training Centre at SMH and CIHR.
Author information
Authors and Affiliations
Contributions
CZ, BZW, and KLT were responsible for the study conception and design, and manuscript preparation. CZ and BZW conducted all functional experiments with assistance from YFW, SSKB, and KLT. CZ, BZW, and KLT performed data analyses. NAP and MST provided PDX tissue samples. AE performed IHC experiments. RC and ISB provided reagents and assisted with custom library design and synthesis. CDCO contributed to microscopy data acquisition and processing. WLL provided clinical data for the BCCA dataset (GSE75037). MRB, TWM and DWC provided CFI-400945 and other essential reagents. All authors made significant contributions and critically reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
Christopher Zhang, Benson Z. Wu, Caterina Di Ciano-Oliveira, Yin Fang Wu, Sharon S. Khavkine Binstock, Nhu-An Pham, Andrew J. Elia, Raj Chari, Wan L. Lam, Ming S. Tsao, Kelsie L. Thu – none. Isabel Soria-Bretones - ISB is currently an employee of Repare Therapeutics. Mark Bray - MRB is currently an employee of Treadwell Therapeutics. Tak Mak - Owns equity in Treadwell Therapeutics Inc. and Agios Pharmaceuticals and is a consultant for AstraZeneca and Tessa Therapeutics. David Cescon - DWC reports consultancy and advisory relationships with AstraZeneca, Daiichi Sankyo, Exact Sciences, Eisai, Gilead, GlaxoSmithKline, Inflex, Inivata/NeoGenomics, Lilly, Merck, Novartis, Pfizer, Roche and Saga; research funding to their institution from AstraZeneca, Guardant Health, Gilead, GlaxoSmithKline, Grail, Inivata/NeoGenomics, Knight, Merck, Pfizer, ProteinQure and Roche.
Ethical approval
Tumor tissues were banked after informed written patient consent following a protocol approved by the University Health Network Research Ethics Board.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, C., Wu, B.Z., Di Ciano-Oliveira, C. et al. Identification of KIFC1 as a putative vulnerability in lung cancers with centrosome amplification. Cancer Gene Ther (2024). https://doi.org/10.1038/s41417-024-00824-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41417-024-00824-1
- Springer Nature America, Inc.