Abstract
Objective
The objectives of this research were to screen the anti-quorum sensing and antibiofilm activity of marine actinobacteria, isolated from several aquatic environments in Indonesia against several pathogenic bacteria, such as Staphylococcus aureus, Bacillus cereus, Enterococcus faecalis, Vibrio cholerae, Salmonella Typhimurium, and Pseudomonas aeruginosa.
Results
Ten out of 40 actinobacteria were found to have anti-quorum sensing activity against wild-type Chromobacterium violaceum (ATCC 12472); however, the validation assay showed that only eight of 10 significantly inhibited the quorum sensing system of Chromobacterium violaceum CV026. The crude actinobacteria extracts inhibited and disrupted biofilm formation produced by pathogens. The highest antibiofilm inhibition was discovered in isolates 11AC (90%), 1AC (90%), CW17 (84%), TB12 (94%), 20PM (85%), CW01 (93%) against Staphylococcus aureus, Bacillus cereus, Enterococcus faecalis, Vibrio cholerae, Salmonella Typhimurium, and Pseudomonas aeruginosa, respectively. The highest biofilm destruction activity was observed for isolate 1AC (77%), 20PM (85%), 16PM (72%), CW01 (73%), 18PM (82%), 16PM (63%) against Staphylococcus aureus, Bacillus cereus, Enterococcus faecalis, Vibrio cholerae, Salmonella Typhimurium, and Pseudomonas aeruginosa, respectively. Actinobacteria isolates demonstrated promising anti-quorum and/or antibiofilm activity, interfering with the biofilm formation of tested pathogens. Appropriate formulations of these extracts could be developed as effective disinfectants, eradicating biofilms in many industries.
Similar content being viewed by others
Introduction
Antimicrobials have been very effective in arresting pathogens’ growth in many circumstances [1, 2]. Unfortunately, rampant usage and poor patient compliance have accelerated antimicrobial resistance in bacteria. Infection of antimicrobial-resistant (AMR) bacteria can be life-threatening. Moreover, AMR bacteria such as S. aureus and S. Typhimurium, can produce biofilms [3, 4]. In illnesses attributed to biofilm-forming bacteria, extracellular matrix developed, increasing their resistance to chemical treatments up to 1000-fold greater than its planktonic form [5].
The uncontrolled growth of biofilm-forming bacteria causes an enormous number of outbreaks and poses global risks by either infection, intoxication, or toxicoinfection. Therefore, there is a growing need to identify new solutions to control biofilm-forming pathogens [6].
Quorum sensing (QS) is the cell-to-cell communication activity of bacteria using autoinducers that regulates virulence factor and biofilm formation [6, 7]. Consequently, inhibiting QS activity might initiate biofilms’ eradication. Without QS activity, bacterial aptitude to protect themselves will decline drastically.
Actinobacteria are fungi-like Gram-positive bacteria, known as the leading producer of secondary metabolites, including novel anti-QS and biofilm inhibitors [8]. However, the research has been only minimally explored. This research aims to screen anti-QS compounds from actinobacteria using wild-type Chromobacterium violaceum and Chromobacterium violaceum CV026 as QS indicators and quantify their antibiofilm activity against biofilm-forming pathogens.
Main text
Methods
Bacterial cultivation
Actinobacteria isolates were retrieved from previous studies [9, 10]. Actinobacteria were cultured on yeast malt extract agar (YMEA; 4 g yeast extract, 10 g malt extract, 4 g glucose, 2% agar) and incubated at 28 °C for 7 days. Wild-type Chromobacterium violaceum (ATCC 12472) and Chromobacterium violaceum CV026 as QS indicators were cultivated on Luria agar (LA; Oxoid) and incubated at 28 °C for 24 h. C. violaceum CV026 is a pigment-negative strain due to mutation in CviI, encoding AHL synthase for autoinducer production. However, in the presence of exogenous AHL, C. violaceum CV026 will express QS-mediated responses leading to pigment production [11]. Biofilm-forming bacteria B. cereus ATCC 14579, S. aureus ATCC 29213, E. faecalis ATCC 33186, P. aeruginosa ATCC 27853, S. Typhimurium, and V. cholerae were cultivated onto LA, incubated at 37 °C for 24 h. These pathogens can initiate biofilm formation via QS activity [3, 4, 12,13,14,15].
Primary screening of anti-quorum sensing activity
Primary screening was carried out using an overlay agar method [16] with modifications. Actinobacteria isolates were inoculated onto YMEA and incubated at 28 °C for 3 days. Wild-type C. violaceum (OD600 = 0.132, 100 µL) was mixed with 2 mL semi-solid (0.75%) LA and poured atop YMEA plates. The plates were incubated at 28 °C for 24 h. Inhibited violacein production around actinobacteria isolates signifies anti-QS activity.
Preparation of crude extract
The extract was obtained using liquid-liquid extraction. Actinobacteria isolate was grown into tryptic soy broth supplemented (Oxoid) with glucose (1% w/v) at 28 °C and 125 rpm for 7 days. The cultures were centrifuged at 7800×g for 15 min. The supernatant was mixed with ethyl acetate (1:1) and shaken at 150 rpm for 24 h. The solvent was collected and evaporated using a rotary evaporator then dried using a vacuum oven. Crude extract was mixed in 1% v/v dimethyl sulfoxide (DMSO) to provide 5, 10, and 20 mg/mL concentrations [17, 18].
Antimicrobial assay
Antimicrobial assays were performed using the agar well diffusion method [19] with modifications. Pathogens (OD600 = 0.132) were spread onto brain heart infusion agar (BHIA, Oxoid). Wells were created and filled with the extract (50 µL, 5 and 10 mg/mL). DMSO (1% v/v) was used as the negative control, and streptomycin (10 mg/mL) was used as the positive control.
Incubation at 37 °C for 24 h revealed clear zones, indicating antibacterial activity.
Secondary screening of anti-quorum sensing activity
Wild-type C. violaceum (OD540 = 0.132) was spread onto BHIA plates. Wells were created and filled with the extract (50 µL, 5 and 10 mg/mL). DMSO (1% v/v) was used as the control [20]. The plates were incubated at 28 °C for 24 h. Inhibited violacein production indicated anti-QS activity [21].
Quantification of antibiofilm activity
The antibiofilm activity was categorized as inhibition or elimination. To detect inhibition activity, pathogens (OD600 = 0.132, 100 µL) cultivated into Brain Heart Infusion Broth supplemented with glucose (2% w/v), and extracts (5 and 10 mg/mL, 100 µL) were transferred to the 96-well microplate. Biofilm inhibition activity was quantified after 24 h. For the elimination activity assay, another 96-well plate with bacterial culture was incubated at 37 °C for 24 h. After biofilms were attached, extract was added and incubated for 24 h. Each pathogenic culture was used as the positive control, while sterile BHIB was used as the negative control.
After incubation, planktonic cells and media were discarded. Adherent cells were rinsed twice with sterile water and stained with crystal violet (0.4% w/v) for 30 min. The microplate was rinsed twice and air-dried for 5 min. Subsequently, 200 µL of ethanol was mixed. Absorbance was determined at 595 nm [22]. Antibiofilm activity was calculated using this equation:
Validation of Quorum sensing inhibition
Chromobacterium violaceum CV026 (OD600 = 0.132, 100 µL) was mixed with 100 µL of extract (final concentration 10 mg/mL) and 1 µL hexanoyl-l-homoserine-lactone (HHL, final concentration 100 mM, Sigma-Aldrich) diluted in acidified ethyl acetate (0.1% v/v acetic acid). Incubation at 28 °C for 24 h followed. Mixture without extract served as a positive control. After incubation, test tubes were centrifuged at 1000 rpm for 15 min. The pellet was mixed with 1 mL DMSO (1%v/v) and centrifuged at 1000 rpm for 15 min. The supernatant’s absorbance was measured at 540 nm [18]. Furthermore, to investigate whether the main compounds in the extract were protein-based, the determination was carried out using proteinase-K. Each extract was treated with proteinase-K (100 µg/mL) for 2 h incubation at 37 °C and subjected to high-temperature treatment at 95 °C for 1 h. The treated extract was used for the validation assay. Total violacein produced by C. violaceum CV026 was compared with the untreated batch [23].
Statistical analysis
Data were compared using a one-way ANOVA based on a confidence level at 95% and Tukey HSD post-hoc analysis.
Examination of biofilm formation by scanning electron microscope (SEM)
Bacillus cereus and Salmonella Typhimurium culture (OD600 = 0.132) were spotted onto sterile cover glass within sterile Petri dishes and incubated at 37 °C for 24 h to form mature biofilms. After incubation, actinobacterial extract (100 µL, 10 mg/mL) was added and re-incubated for 24 h. The results were investigated using SEM at Dexa Laboratory of Biomolecular and Science [24].
Results
First screening of anti-quorum sensing activity
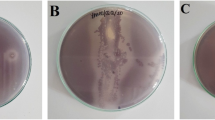
The results show that 10 out of 40 actinobacteria isolates had anti-QS activity (Additional file 1: Illustration S1). Those isolates were extracted and continued to the next step (Additional file 1: Illustration S2).
Antibacterial activity assay
Ten actinobacterial extracts showed no antibacterial activity against S. aureus, B. cereus, E. faecalis, P. aeruginosa, and S. Typhimurium, but 3 of the 10 showed antibacterial activity against V. cholerae. Those extracts were 15PM, 18PM, and 20PM. Streptomycin as the positive control inhibited the growth of all pathogens. In contrast, DMSO as the negative control showed no antibacterial activity (Additional file 1: Illustration S3).
Secondary screening of anti-quorum sensing activity
Ten actinobacterial extracts in 5 and 10 mg/mL concentrations still had anti-QS activity. It was demonstrated by the absence of violacein pigment around the wells (Additional file 1: Illustration S4).
Anti-quorum sensing activity validation test and protein characterization
All treatments yielded lower absorbance than the control (Fig. 1). However, only 8 out of 10 extracts (10 mg/mL) demonstrated statistically significant anti-QS activity against C. violaceum CV026: 1AC, 11AC, 14PM, 15PM, 16PM, 18PM, CW01, and CW17. Treated extract of all isolates showed anti-QS activity distinct from the control. Increased absorbance suggests interference with anti-QS compounds due to proteolytic and high-temperature treatments, while lower values indicate heightened anti-QS activity.
Biofilm inhibition and elimination assay
Crude extracts showed diverse antibiofilm activity in inhibiting and eliminating bacterial biofilms (Table 1).
Examination of biofilm formation by scanning electron microscope
The result showed the topographical of B. cereus and S. Typhimurium biofilms before and after treatment with actinobacterial extract. Considerable morphological changes occurred in bacterial biofilms.
Discussion
The primary screening of anti-QS activity showed that 10 out of 40 isolates demonstrated anti-QS activity. In adapting to competitive environments, actinobacteria produce secondary metabolites such as anti-QS and anti-biofilms [25]. No clear zones were observed in the antimicrobial assay, indicating negative-antimicrobial activity, except in V. cholerae. Of the 10 actinobacteria isolates, three (15PM, 18PM, and 20PM) inhibited the growth of V. cholerae. Those isolates were excluded in the V. cholerae antibiofilm assay. Extract in 5 and 10 mg/mL concentrations inhibited the QS system of wild-type C. violaceum, as seen from lower pigment production around the wells. From the validation assay, only 8 of 10 isolates inhibited the QS activity of C. violaceum CV026. The compounds in the extract might be damaging other components of the QS system, such as the stability and function of AHL, autoinducer synthase, and its regulators [26, 27].
Anti-QS agents in extracts of 1AC, 14PM, 16PM, 18PM, and CW17 isolates were likely proteins because higher absorbance value observed after proteinase-K and high-temperature treatment. The increasing absorbance value implied reduced anti-QS activity. The treatment might affect proteinaceous anti-QS compounds produced by actinobacteria, such as AHL-lactonase, acylase, decarboxylase, oxidoreductase, and deaminase [28], which the treatment can easily degrade. In the absence of anti-QS compounds, AHL will form protein-ligand complex, activating violacein production [29].
Extract from isolates 15PM, 20PM, CW01, and TB12 treated with proteinase-K showed higher anti-QS activity, indicating degradation of proteins that competitively bind to similar receptors or any proteins that might inhibit the anti-QS activity [30]. Additionally, anti-QS compounds might be a proenzyme or peptides hence the reaction with proteinase-K would positively affect the anti-QS activity. LaSarre reported actinobacteria produce proteinaceous compounds activated by serine proteases, enhancing anti-QS activity. [3, 8, 31].
Crude extracts had produced without fractionation and purification processes, potentially containing inhibitors or analogs that affect compound interactions. The interactions between undesirable compounds inevitably impact the effectiveness [32]. It might work antagonistically or synergistically, which could decrease or increase inhibition activity [31,32,33].
Biofilms consist of microorganisms and extracellular polymeric substances (EPSs). Disrupting the EPS layer might severely reduce biofilm formation. In S. aureus, the primary EPS components are polysaccharide intercellular adhesin, known as poly-N-acetyl-β-(1–6)-glucosamine, polysaccharide, proteins, and extracellular DNA (eDNA) [34]. B. cereus utilizes exopolysaccharides, proteins, and eDNA [35]. The EPSs of E. faecalis are predominantly eDNA and polysaccharides [36]. While P. aeruginosa’s EPS comprises eDNA, proteins, lipids, and polysaccharides, such as Psl, Pel, and alginate [37]. In S. Typhimurium, curli fimbriae and cellulose drive cell clustering and biofilm initiation [38]. In V. cholerae biofilms consist of vibrio-exopolysaccharides, proteins, and eDNA [39]. Based on previous researches [9, 40], Most of the isolates are belonging to the genera Streptomyces and Arthrobacter. Streptomyces produces ECM-degrading enzymes such as protease, amylase, nuclease, agarase, chitinase, and several organic acids such as docosanoic acid, tetracosanoic acid, arachidic acid, and erucic acid that are capable of inhibiting P. aeruginosa and S. aureus biofilms [41, 42]. Arthrobacter was reported to produce dextranase and xylanase, which may significantly eradicate the P. aeruginosa, methicillin-resistance S. aureus, and E. coli biofilms. Additionally, Arthrobacter produces glucuronide and cyclic-depsipeptides, such as arthroamide and turnagainolide, that could interfere the QS system [42, 43].
Scanning electron microscopy showed the presence of B. cereus and S. Typhimurium biofilms. Antibiofilm activity can be observed due to the depletion and erosion across biofilm surfaces. Both extracts were competent to degrade the biofilm formation (Fig. 2) [44].
Conclusions
In summary, this study showed the potential of actinobacterial extracts performing anti-QS activity against C. violaceum along with biofilm inhibition and elimination activities against biofilm-forming pathogens. They could be developed as a safer and more efficient disinfectants in several industries.
Limitations
We did not determine the specific compound(s) responsible for antibiofilm and anti-QS activity and each isolate’s anti-QS mechanism. Furthermore, our characterization was limited to protein.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AHL:
-
Acyl homoserine lactone
- AMR:
-
Antimicrobial resistance
- ANOVA:
-
Analysis of variance
- BHIA:
-
Brain heart infusion agar
- BHIB:
-
Brain heart infusion broth
- ECM:
-
Extracellular matrix
- eDNA:
-
Extracellular DNA
- EPS:
-
Extra polymeric substances
- HHL:
-
Hexanoyl-l-homoserine-lactone
- LA:
-
Luria agar
- LB:
-
Luria broth
- QS:
-
Quorum sensing
- SEM:
-
Scanning electron microscope
- TSB:
-
Tryptone soy broth
- YMEA:
-
Yeast malt extract agar
References
Aminov RI. A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol. 2010;134:1–7.
Gaynes R. The discovery of penicillin—new insights after more than 75 years of clinical use. Emerg Infect Dis. 2017;23:849–53.
Gkana EN, Giaouris ED, Doulgeraki AI, Kathariou S, Nychas GJE. Biofilm formation by Salmonella Typhimurium and Staphylococcus aureus on stainless steel under either mono- or dual-species multi-strain conditions and resistance of sessile communities to sub-lethal chemical disinfection. Food Control. 2017;73:838–46.
Neopane P, Nepal HP, Shrestha R, Uehara O, Abiko Y. In vitro biofilm formation by Staphylococcus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance. Int J Gen Med. 2018;11:25–32.
Mah T-F, Pitts B, Pellock B, Walker GC, Stewart PS, O’Toole GA. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–10.
Rabin N, Zheng Y, Opoku-Temeng C, Du Y, Bonsu E, Sintim HO. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med Chem. 2015;7:493–512.
Das S, Dash HR. Microbial biotechnology—a laboratory manual for bacterial systems. New Delhi: Springer India; 2015.
Chater KF, Biró S, Lee KJ, Palmer T, Schrempf H. The complex extracellular biology of Streptomyces. FEMS Microbiol Rev. 2010;34:171–98.
Andreas. Screening of actinomycetes from marine sediments to inhibit Vibrio cholerae biofilm formation. [Biotechnology Thesis]. Jakarta: Atma Jaya Catholic University of Indonesia; 2011.
Vidyawan V. Screening of actinomycetes from various environment sediments to inhibit biofilm formation of Vibrio cholerae [Biotechnology Thesis]. Jakarta: Atma Jaya Catholic University of Indonesia; 2012.
Kothari V, Sharma S, Padia D. Recent research advances on Chromobacterium violaceum. Asian Pac J Trop. 2017;10:744–52.
Di Ciccio PA. Antimicrobial-resistance of foodborne pathogens. Antibiotics. 2021;10: 372.
Fernández-Delgado M, Rojas H, Duque Z, Suárez P, Contreras M, García-Amado MA, Alciaturi C. Biofilm formation of Vibrio cholerae on stainless steel used in food processing. Rev Inst Med Trop S Paulo. 2016. https://doi.org/10.1590/S1678-9946201658047.
Kaverimanian V, Heuertz RM. Effects of neem extracts on formed biofilm of Pseudomonas aeruginosa. FASEB J. 2020;34:1–1.
Kim M-A, Rosa V, Min K-S. Characterization of Enterococcus faecalis in different culture conditions. Sci Rep. 2020;10:21867.
Abudoleh SM, Mahasneh AM. Anti-quorum sensing activity of substances isolated from wild berry associated bacteria. Avicenna J Med Biotechnol. 2017;9:23–30.
Nithya C, Aravindraja C, Pandian SK. Bacillus pumilus of Palk Bay origin inhibits quorum-sensing-mediated virulence factors in Gram-negative bacteria. Res Microbiol. 2010;161:293–304.
Rajivgandhi G, Senthil R, Ramachandran G, Maruthupandy M, Manoharan N. Antibiofilm activity of marine endophytic actinomycetes compound isolated from mangrove plant Rhizophora mucronata, Muthupet Mangrove Region, Tamil Nadu, India. J Terr Mar Res. 2018;2:7.
Nathan P, Law EJ, Murphy DF, MacMillan BG. A laboratory method for selection of topical antimicrobial agents to treat infected burn wounds. Burns. 1978;4:177–87.
Rajivgandhi G, Vijayan R, Maruthupandy M, Vaseeharan B, Manoharan N. Antibiofilm effect of Nocardiopsis sp. GRG 1 (KT235640) compound against biofilm forming Gram negative bacteria on UTIs. Microb Pathog. 2018;118:190–8.
Khan MSA, Zahin M, Hasan S, Husain FM, Ahmad I. Inhibition of quorum sensing regulated bacterial functions by plant essential oils with special reference to clove oil. Lett Appl Microbiol. 2009;49:354–60.
Waturangi DE, Purwa HJ, Lois W, Hutagalung RA, Hwang JK. Inhibition of marine biofouling by aquatic actinobacteria and coral-associated marine bacteria. Malays J Microbiol. 2017. https://doi.org/10.21161/mjm.86016.
Thenmozhi R, Nithyanand P, Rathna J, Karutha Pandian S. Antibiofilm activity of coral-associated bacteria against different clinical M serotypes of Streptococcus pyogenes. FEMS Immunol Med Microbiol. 2009;57:284–94.
Luo J, Dong B, Wang K, Cai S, Liu T, Cheng X, et al. Baicalin inhibits biofilm formation, attenuates the quorum sensing-controlled virulence and enhances Pseudomonas aeruginosa clearance in a mouse peritoneal implant Infection model. PLoS ONE. 2017;12: e0176883.
Amara N, Krom BP, Kaufmann GF, Meijler MM. Macromolecular inhibition of quorum sensing: enzymes, antibodies, and beyond. Chem Rev. 2011;111:195–208.
Martinelli D, Grossmann G, Séquin U, Brandl H, Bachofen R. Effects of natural and chemically synthesized furanones on quorum sensing in Chromobacterium violaceum. BMC Microbiol. 2004;4: 25.
Vasavi HS, Arun AB, Rekha PD. Inhibition of quorum sensing in Chromobacterium violaceum by Syzygium cumini L. and Pimenta dioica L. Asian Pac J Trop Biomed. 2013;3:954–9.
Chen F, Gao Y, Chen X, Yu Z, Li X. Quorum quenching enzymes and their application in degrading signal molecules to block quorum sensing-dependent Infection. Int J Mol Sci. 2013;14:17477–500.
Stauff DL, Bassler BL. Quorum sensing in Chromobacterium violaceum: DNA recognition and gene regulation by the CviR receptor. J Bacteriol. 2011;193:3871–8.
Aljelawi RO, Kadhem MF. Production, purification, and characterization of bioactive metabolites produced from rare actinobacteria Pseudonocardia alni. Asian J Pharm Clin Res. 2016;9:264.
LaSarre B, Federle MJ. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol Mol Biol Rev. 2013;77:73–111.
Caesar LK, Cech NB. Synergy and antagonism in natural product extracts: when 1 + 1 does not equal 2. Nat Prod Rep. 2019;36:869–88.
Giguère S, Lee EA, Guldbech KM, Berghaus LJ. In vitro synergy, pharmacodynamics, and postantibiotic effect of 11 antimicrobial agents against Rhodococcus equi. Vet Microbiol. 2012;160:207–13.
Polkade AV, Mantri SS, Patwekar UJ, Jangid K. Quorum sensing: an under-explored phenomenon in the phylum actinobacteria. Front Microbiol. 2016. https://doi.org/10.3389/fmicb.2016.0013133.
Majed R, Faille C, Kallassy M, Gohar M. Bacillus cereus biofilms—same, only different. Front Microbiol. 2016. https://doi.org/10.3389/fmicb.2016.01054.
Chen W, Liang J, He Z, Jiang W. Differences in the chemical composition of Enterococcus faecalis biofilm under conditions of Starvation and alkalinity. Bioengineered. 2017;8:1–7.
Thi MTT, Wibowo D, Rehm BHA. Pseudomonas aeruginosa biofilms. Int J Mol Sci. 2020;21: 8671.
Römling U, Bokranz W, Rabsch W, Zogaj X, Nimtz M, Tschäpe H. Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human Disease. Int J Med Microbiol. 2003;293:273–85.
Waturangi DE, Rahayu BS, Lalu MKY, Mulyono N. Characterization of bioactive compound from actinomycetes from antibiofilm activity against Gram-negative and Gram-positive bacteria. Malays J Microbiol. 2016. https://doi.org/10.21161/mjm.80915.
Silva AJ, Benitez JA. Vibrio cholerae biofilms and cholera pathogenesis. PLoS Negl Trop Dis. 2016;10: e0004330.
Kamarudheen N, Rao KVB. Fatty acyl compounds from marine Streptomyces griseoincarnatus strain HK12 against two major biofilm forming nosocomial pathogens; an in vitro and in silico approach. Microb Pathog. 2019;127:121–30.
Goel N, Fatima SW, Kumar S, Sinha R, Khare SK. Antimicrobial resistance in biofilms: exploring marine actinobacteria as a potential source of antibiotics and biofilm inhibitors. Biotechnol Rep. 2021;30:e00613.
Sarveswari HB, Solomon AP. Profile of the intervention potential of the phylum actinobacteria toward quorum sensing and other microbial virulence strategies. Front Microbiol. 2019;10: 2073.
Li XH, Lee JH. Antibiofilm agents: a new perspective for antimicrobial strategy. J Microbiol. 2017;55:753–66.
Acknowledgements
The authors thankfully acknowledge the microscopy facility provided by the PT Dexa Laboratories of Biomolecular Sciences (DLBS).
Funding
This research had funded by the Ministry of Research, Technology and Higher Education of the Republic of Indonesia 2019 and Faculty of Biotechnology, Atma Jaya Catholic University of Indonesia 2019. The funder has no contribution in the design, data collection, writing, and data interpretation in this research.
Author information
Authors and Affiliations
Contributions
Material preparation, data collection and analysis were performed by MW and DD under the advisory of DEW. DEW conception design and data analysis. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Illustration S1.
Primary screening of anti-quorum sensing activity performed by 18PM Isolates against wild-type C. violaceum (ATCC 12472). Illustration S2. Ten actinobacteria isolates with positive anti-quorum sensing against wild-type C. violaceum (ATCC 12472). Illustration S3. Antimicrobial assay of actinobacterial crude extracts (50 μL, 10 mg/mL) against tested bacteria (a) S. aureus (b) E. faecalis (c) B. cereus (d) V. cholerae (e) S. Typhimurium (f) P. aeruginosa with K+: streptomycin (20 μL;10 mg/mL) as positive control and K-: DMSO (50 μL; 1%v/v) as negative control. Illustration S4. Secondary screening of actinobacterial crude extract of 18PM and 20PM (50 μL) (a) 5 mg/mL and (b) 10 mg/mL against wild-type C. violaceum (ATCC 12472) with K−: DMSO (50 μL; 1%v/v) as negative control.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wijaya, M., Delicia, D. & Waturangi, D.E. Control of pathogenic bacteria using marine actinobacterial extract with antiquorum sensing and antibiofilm activity. BMC Res Notes 16, 305 (2023). https://doi.org/10.1186/s13104-023-06580-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13104-023-06580-z